For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSEVKQHWEERYAERDRIWSGRVNRWLAEVAASEKPGHALDLGCGEGGDTVWLAEHGWQVLGVDISDTAL GRAAAEAATRGLTDRVRFVQMNLSEAFPEGVFDLVSAQFLQSMVHLDRERIFAAAARAVAPGGVLVIVDH GAAPSWAPEHIREHVFPGVEEVLATVDLGDGQWRRVRADRVERDATGPDGQQAVLIDNLIVLRRL*
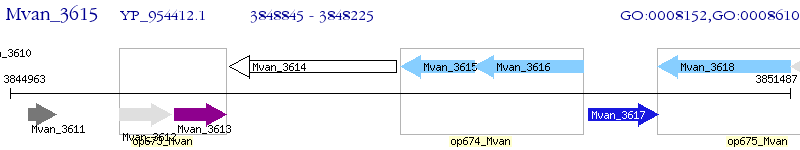
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3615 | - | - | 100% (206) | methyltransferase type 11 |
| M. vanbaalenii PYR-1 | Mvan_5314 | - | 1e-19 | 34.30% (207) | methyltransferase type 11 |
| M. vanbaalenii PYR-1 | Mvan_4027 | - | 6e-12 | 31.85% (135) | methyltransferase type 12 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3351c | - | 6e-12 | 40.18% (112) | methyltransferase |
| M. gilvum PYR-GCK | Mflv_2895 | - | 6e-90 | 76.35% (203) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv3322c | - | 6e-12 | 40.18% (112) | methyltransferase |
| M. leprae Br4923 | MLBr_01713 | - | 8e-05 | 24.29% (177) | hypothetical protein MLBr_01713 |
| M. abscessus ATCC 19977 | MAB_3178 | - | 2e-29 | 47.10% (138) | hypothetical protein MAB_3178 |
| M. marinum M | MMAR_2640 | - | 5e-12 | 33.81% (139) | methyltransferase |
| M. avium 104 | MAV_2216 | cheR | 2e-60 | 56.44% (202) | CheR methyltransferase SAM binding domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_4303 | - | 5e-66 | 60.30% (199) | methyltransferase |
| M. thermoresistible (build 8) | TH_0144 | - | 3e-62 | 61.50% (187) | methyltransferase |
| M. ulcerans Agy99 | MUL_3115 | - | 2e-13 | 33.33% (138) | methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3351c|M.bovis_AF2122/97 --------------------------------------------------
Rv3322c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2640|M.marinum_M --------------------------------------------------
MUL_3115|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3615|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2895|M.gilvum_PYR-GCK --------------------------------------------------
MAV_2216|M.avium_104 --------------------------------------------------
TH_0144|M.thermoresistible__bu --------------------------------------------------
MSMEG_4303|M.smegmatis_MC2_155 MENKPWDSIIVGGGAAGLSAALVLGRARRRVLLVDAGEQSNLPAHGIGGL
MAB_3178|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01713|M.leprae_Br4923 --------------------------------------------------
Mb3351c|M.bovis_AF2122/97 --------------------------------------------------
Rv3322c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2640|M.marinum_M --------------------------------------------------
MUL_3115|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3615|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2895|M.gilvum_PYR-GCK --------------------------------------------------
MAV_2216|M.avium_104 --------------------------------------------------
TH_0144|M.thermoresistible__bu --------------------------------------------------
MSMEG_4303|M.smegmatis_MC2_155 LGSDGRAPADLYAAGHAELAAYPSVQVRRGEVIAGRRGFALDLADGGHEH
MAB_3178|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01713|M.leprae_Br4923 --------------------------------------------------
Mb3351c|M.bovis_AF2122/97 --------------------------------------------------
Rv3322c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2640|M.marinum_M --------------------------------------------------
MUL_3115|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3615|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2895|M.gilvum_PYR-GCK --------------------------------------------------
MAV_2216|M.avium_104 --------------------------------------------------
TH_0144|M.thermoresistible__bu --------------------------------------------------
MSMEG_4303|M.smegmatis_MC2_155 ARTVLLAAGMDYLPPNIPGLGELWGRSVFHCPFCHGWEVRDAPLAVIARG
MAB_3178|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01713|M.leprae_Br4923 --------------------------------------------------
Mb3351c|M.bovis_AF2122/97 --------------------------------------------------
Rv3322c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2640|M.marinum_M --------------------------------------------------
MUL_3115|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3615|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2895|M.gilvum_PYR-GCK --------------------------------------------------
MAV_2216|M.avium_104 --------------------------------------------------
TH_0144|M.thermoresistible__bu --------------------------------------------------
MSMEG_4303|M.smegmatis_MC2_155 ERAVHSALLLRAWSDDIVVLTNGPDGLDDAQTEKLGAAGIPVNDASIARL
MAB_3178|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01713|M.leprae_Br4923 --------------------------------------------------
Mb3351c|M.bovis_AF2122/97 --------------------------------------------------
Rv3322c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2640|M.marinum_M --------------------------------------------------
MUL_3115|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3615|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2895|M.gilvum_PYR-GCK --------------------------------------------------
MAV_2216|M.avium_104 --------------------------------------------------
TH_0144|M.thermoresistible__bu --------------------------------------------------
MSMEG_4303|M.smegmatis_MC2_155 VAHDGELVAVEFTDGTRLSRRGALVATTLHQRSPLAAQLGAIGAPTPLAG
MAB_3178|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01713|M.leprae_Br4923 --------------------------------------------------
Mb3351c|M.bovis_AF2122/97 --------------------------------------------------
Rv3322c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2640|M.marinum_M -------------------------MSRTAERADRLSIRYPVCSPTAQVR
MUL_3115|M.ulcerans_Agy99 --------------------------------------------------
Mvan_3615|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2895|M.gilvum_PYR-GCK --------------------------------------------------
MAV_2216|M.avium_104 --------------------------------------------------
TH_0144|M.thermoresistible__bu --------------------------------------------------
MSMEG_4303|M.smegmatis_MC2_155 DAVVVDTFGRTTVPGLFAAGDLTGNLPQVATAVAAGSQTAAAIVQHLLAE
MAB_3178|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01713|M.leprae_Br4923 --------------------------------------MSAFVPDVPNRI
Mb3351c|M.bovis_AF2122/97 --MSVQTDPALREHPNRVDWNARYERAG-SAHAPFAPVPWLADVLRAGVP
Rv3322c|M.tuberculosis_H37Rv --MSVQTDPALREHPNRVDWNARYERAG-SAHAPFAPVPWLADVLRAGVP
MMAR_2640|M.marinum_M RSCRGRRMVARVAESDRRRWDRRYASLGPAAVSAARPPALLQAHLKLVPT
MUL_3115|M.ulcerans_Agy99 --------MARVAESDRRRWDRRYASLGPAAVSAVRPPALLQAHLKLVPA
Mvan_3615|M.vanbaalenii_PYR-1 ---MTS--------EVKQHWEERYAERD--RIWSGRVNRWLAEVAASEKP
Mflv_2895|M.gilvum_PYR-GCK ---MT---------DVKQHWEERYSQRD--RIWSGHVNRWLAEVAAPLKP
MAV_2216|M.avium_104 ---MDTSCEPGE---AGRFWEERYRGAE--RVWSGRVNPRLAELAADLPA
TH_0144|M.thermoresistible__bu -------------------------------MWSGRVNARLAEVAGELPP
MSMEG_4303|M.smegmatis_MC2_155 DIGLPTPPWPTEGLSPAEYWERHYGERE--QIWSGRVNPQLEKVASDLPA
MAB_3178|M.abscessus_ATCC_1997 ---MHDHNNPPS---DEKSWDEFYQSHD--ALWSGNANPQLVAEISSLPP
MLBr_01713|M.leprae_Br4923 PDDPSSIDSTAFHGMLTLTGERTIPDLDIENYWFRRHQVVYQWLAQRCTG
Mb3351c|M.bovis_AF2122/97 DGPVLELASGRSGTALALAAHGRQVTAIDVSDVALLQLDSEAVRR--GVA
Rv3322c|M.tuberculosis_H37Rv DGPVLELASGRSGTALALAAHGRQVTAIDVSDVALLQLDSEAVRR--GVA
MMAR_2640|M.marinum_M AGRALDLGCGQGLGAVWLARRGLDVLGLDISPVAIAQARDLAERS--GVS
MUL_3115|M.ulcerans_Agy99 AGRALDLGCGQGLGAVWLARRGLDVLGLDISPVAIAQARDLAERS--GVS
Mvan_3615|M.vanbaalenii_PYR-1 -GHALDLGCGEGGDTVWLAEHGWQVLGVDISDTALGRAAAEAATR--GLT
Mflv_2895|M.gilvum_PYR-GCK -GRALDLGCGEGGDTLWLAEHGWEVVGVDISDTALRRAAEEADKR--GLA
MAV_2216|M.avium_104 -GRALDLGCGEGADALWLAQRGWTVLAVDISATALRRAAEAAASR--KLL
TH_0144|M.thermoresistible__bu -GRALDVGCGEGADAMWLAERGWQVTAVDISQTALDRAAAEAASR--GLT
MSMEG_4303|M.smegmatis_MC2_155 -GHALDLGCGEGGDAVWLAERGWRVTAVDISHTALSRAAAEAGRR--GVA
MAB_3178|M.abscessus_ATCC_1997 -RTALDAGCGEGGDAIWLAQHGWQVTGMDFAHTALRRAADNAVKVDPELA
MLBr_01713|M.leprae_Br4923 -CEVLEAGCGEGYGADLIVDVAYRVIAVDYDEATVAHVRSRYPRV---QV
.*: ..*.. : :. . * .:* .:: :
Mb3351c|M.bovis_AF2122/97 DRLNLVQADLGCWEPGETR-FALVLSRLFW------DAAIFHRACEAVMP
Rv3322c|M.tuberculosis_H37Rv DRLNLVQADLGCWEPGETR-FALVLSRLFW------DAAIFHRACEAVMP
MMAR_2640|M.marinum_M ARCCFDVVDLDDGLPDGPP-VDVIVCHNFR------DRRLDAAIIDRLAP
MUL_3115|M.ulcerans_Agy99 ARCRFDVVDLDDGLPDGPP-VDLIVCHNFR------DRRLDAAIIERLAP
Mvan_3615|M.vanbaalenii_PYR-1 DRVRFVQMNLSEAFPEGV--FDLVSAQFLQSMVHLDRERIFAAAARAVAP
Mflv_2895|M.gilvum_PYR-GCK DRVQLRQTDLSASFPDGV--FDLVSAQFLQSMVHLDRERIFAAAARAVAP
MAV_2216|M.avium_104 ARIDFQRHDLNESFPEGM--FDLISAQYFHSPVHLDRDAVLRRAATRVKP
TH_0144|M.thermoresistible__bu --IDFQRHELPATFPDGQ--FELVSAHFLHSEVELDRVRILRRAAEAVVP
MSMEG_4303|M.smegmatis_MC2_155 DRITFERHDLSESFPDGE--FDLVSAQFLHSTVRLERPRILSTAARAIRP
MAB_3178|M.abscessus_ATCC_1997 NRISWTQADLTAWQPNRQ--FDLVTSHFMHLPTNL-REPVFAALAAAVAP
MLBr_01713|M.leprae_Br4923 MQANLIELPLADASVDVVVNFQVIEHLWNQAQFVSECARVLRPSGLLMVS
* . :: : : .
Mb3351c|M.bovis_AF2122/97 GGVLAWESLALSG------------AEAGTASAKRRVKPGEPACLLPADF
Rv3322c|M.tuberculosis_H37Rv GGVLAWESLALSG------------AEAGTASAKRRVKPGEPACLLPADF
MMAR_2640|M.marinum_M GGLVALVALSEVG------------AAAGTY----RAAPGELSAAF-AEL
MUL_3115|M.ulcerans_Agy99 GGLVALVALSEVG------------AAAGTY----RAAPGELSTAF-AEL
Mvan_3615|M.vanbaalenii_PYR-1 GGVLVIVDHGAAPSWAPEHIREHVFPGVEEVLATVDLGDGQWRRVRADRV
Mflv_2895|M.gilvum_PYR-GCK GGVLIVVDHAAAPPWAPPEVHEHRFSTVDEVLATIDLGEGSWERLRADVV
MAV_2216|M.avium_104 GGVLLIVDHGAAPPWAQHDGHH--FPGVEEVLGSLRLDPDGWTRLRAEAA
TH_0144|M.thermoresistible__bu GGLLLIVDHGAPPPWAPDHVHEFEFPSAQEVVESLQLPEGQWELLRAEPV
MSMEG_4303|M.smegmatis_MC2_155 GGHLVIVDHGKMPPNATRVPHDHPFPSAEEVLAGLDLPASEWERVRVEAV
MAB_3178|M.abscessus_ATCC_1997 GGTLLIVGHHPSDMQAAIGRPDLPDWYFTAEDIADTLNPKEWTIIVAESR
MLBr_01713|M.leprae_Br4923 TPNRITFSPGRDTSINPFHTRELNADELTQLLITADFSEVAMYGLFHGPR
Mb3351c|M.bovis_AF2122/97 TVVHEGQGNCDSAPSRIMIARRSPLPGA----------------------
Rv3322c|M.tuberculosis_H37Rv TVVHEGQGNCDSAPSRIMIARRSPLPGA----------------------
MMAR_2640|M.marinum_M DPIAAGEG-----DGRAWLLARA---------------------------
MUL_3115|M.ulcerans_Agy99 DPIAAGEG-----DGRAWLLARA---------------------------
Mvan_3615|M.vanbaalenii_PYR-1 ERDATGPDGQQAVLIDNLIVLRRL--------------------------
Mflv_2895|M.gilvum_PYR-GCK ERDAIGPDGEAGVLRDNLIVVRRCA-------------------------
MAV_2216|M.avium_104 GREMTGPNGEVGTLMDNIMMLRRTEQAYAAS-------------------
TH_0144|M.thermoresistible__bu KRDAVGPDGQQGTLIDNVIMLRRR--------------------------
MSMEG_4303|M.smegmatis_MC2_155 HREATGPDGQPMPMVDNVMVLRRTS-------------------------
MAB_3178|M.abscessus_ATCC_1997 SRKFTR-DDEEFDIADTVLRAERKPTTESA--------------------
MLBr_01713|M.leprae_Br4923 LKEMDARHGGSIIDAQIARAVSNAPWPPELVADVAAVTAADFDIIEASAG
Mb3351c|M.bovis_AF2122/97 ----------------------
Rv3322c|M.tuberculosis_H37Rv ----------------------
MMAR_2640|M.marinum_M ----------------------
MUL_3115|M.ulcerans_Agy99 ----------------------
Mvan_3615|M.vanbaalenii_PYR-1 ----------------------
Mflv_2895|M.gilvum_PYR-GCK ----------------------
MAV_2216|M.avium_104 ----------------------
TH_0144|M.thermoresistible__bu ----------------------
MSMEG_4303|M.smegmatis_MC2_155 ----------------------
MAB_3178|M.abscessus_ATCC_1997 ----------------------
MLBr_01713|M.leprae_Br4923 PEASDGRDVDESLDLIAIAVRR