For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ELRQLEYLVAVVEERSFTKAAQRERVAQPAVSAQIRRLERQVGQPLLTRSHRGVRVTPAGAALLPHARAA LAAVRDAQAAVDDVANLVRGAVAIGTVTLHPVDVARLMADFHAEHPGVEITLGTDTSDVLLAKLEDGRLD AAIVSIGADEDPAGLEYEVITDEALEAAVTAQHRLARRKSLSLKQLCEYPVISLPAGTGLRSRLDDACAA AGLRPRIAFEATSPLELADLARHGLGVAILPRSMARGSAGLHPLRLSPELRGRLVWAWRTDISGPAARLL NARARAMIAATGTPAVIPGA*
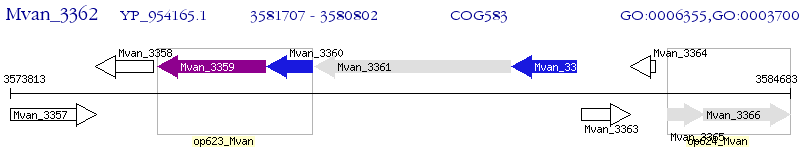
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3362 | - | - | 100% (301) | LysR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0328 | - | 4e-37 | 35.29% (289) | LysR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_4734 | - | 2e-25 | 37.67% (223) | LysR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0121 | oxyS | 6e-17 | 26.23% (305) | oxidative stress response regulatory protein OXYS |
| M. gilvum PYR-GCK | Mflv_3565 | - | 1e-124 | 76.24% (303) | LysR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0117 | oxyS | 6e-17 | 26.23% (305) | oxidative stress response regulatory protein OXYS |
| M. leprae Br4923 | MLBr_02041 | oxyR | 1e-22 | 30.86% (243) | putative LysR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_2238 | - | 5e-73 | 50.34% (290) | LysR family transcriptional regulator |
| M. marinum M | MMAR_4157 | - | 1e-24 | 32.26% (248) | LuxR family transcriptional regulator |
| M. avium 104 | MAV_2838 | - | 2e-27 | 33.61% (241) | hydrogen peroxide-inducible genes activator |
| M. smegmatis MC2 155 | MSMEG_3840 | - | 4e-85 | 57.14% (301) | LysR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_3504 | - | 1e-15 | 31.51% (219) | PUTATIVE putative transcriptional regulator |
| M. ulcerans Agy99 | MUL_4020 | - | 3e-24 | 32.26% (248) | LuxR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_3362|M.vanbaalenii_PYR-1 -------MELRQLEYLVAVVEERSFTKAAQRERVAQPAVSAQIRRLERQV
Mflv_3565|M.gilvum_PYR-GCK -------MELRQLEYFVAIVEEGGFTRAASRERVAQPAVSAQIRRLERSV
MSMEG_3840|M.smegmatis_MC2_155 -------MELRQLEYFVAVAEEANFTRAAQRIHVAQPAVSASIRQLEREL
MAB_2238|M.abscessus_ATCC_1997 -------MEIHQLRYLLAVADTSSFTKAAAALHVAQPGVSAQIRLLEREL
MMAR_4157|M.marinum_M -------MELYQLRYFQAVAECGTLRGASEQLLVSQSAVSRAITLLEAEI
MUL_4020|M.ulcerans_Agy99 -------MELYQLRYFQAVAECGTLRGASEQLLVSQSAVSRAITLLEAEI
MLBr_02041|M.leprae_Br4923 MSDKSYQPTIAGLRAFVAVVEKGHFSAAASFLGVRQSTLSQALAALESGL
MAV_2838|M.avium_104 MPDKTYQPTIAGLRAFVAVAEKRQFSGAATALGVSQSTLSQALAALEAGL
Mb0121|M.bovis_AF2122/97 -------MLFRQLEYFVAVAQERHFARAAEKCYVSQPALSSAIAKLEREL
Rv0117|M.tuberculosis_H37Rv -------MLFRQLEYFVAVAQERHFARAAEKCYVSQPALSSAIAKLEREL
TH_3504|M.thermoresistible__bu --VPSPKIDLRRLAQFVAVAELGSFTRAAATLHLSQQALSSAVRALENDV
: * : *:.: : *: : * :* : ** :
Mvan_3362|M.vanbaalenii_PYR-1 GQPLLTRSHRGVRVTPAGAALLPHARAALAAVRDAQAAVDDVANLVRGAV
Mflv_3565|M.gilvum_PYR-GCK GQPLLTRTSREVRLTRAGEALLPHARAALAAVRDGRAAVDDVAGLLRGSV
MSMEG_3840|M.smegmatis_MC2_155 GQPLFDRSRREVRLTAAGEAVLPHAHAALRSVAAARTAVAEVSGLVRGSV
MAB_2238|M.abscessus_ATCC_1997 GQDLFDRSGRQVRLTDAGKAVIPYARAALAAVDGVREAADEIGGLIRGHI
MMAR_4157|M.marinum_M GVQLFTRRGRTNELNRYGQALLVGSRLAQRSVSSAIDNVRELAGVSGGTV
MUL_4020|M.ulcerans_Agy99 GVQLFTRRGRTNELNRYGQALLVGSRLAQRSVSSAIDNVRELAGVSGGTV
MLBr_02041|M.leprae_Br4923 GVQLIERSTRRVFVTTQGRHLLPRAQAAIEAMDAFTAAAAGESDPLRGGM
MAV_2838|M.avium_104 GTQLVERSTRRVFLTPQGAELLPHAQAVVEAADAFTAAAAGSADPLRAGM
Mb0121|M.bovis_AF2122/97 NVTLINRGHSFEGLTREGERLVVWAKRILAEHAAFKAEVDAVRSGITGTL
Rv0117|M.tuberculosis_H37Rv NVTLINRGHSFEGLTREGERLVVWAKRILAEHAAFKAEVDAVRSGITGTL
TH_3504|M.thermoresistible__bu GADLFTRDGRRISLTTAGRTLLAEGKPLLAAARTVAEHVRQTEAAAQAHY
. *. * :. * :: .: . .
Mvan_3362|M.vanbaalenii_PYR-1 AIGTVTLHPVD-VARLMADFHAEHPGVEITLGT-DTSDVLLAKLEDGRLD
Mflv_3565|M.gilvum_PYR-GCK AIGTVTLHPMN-VAHLIAEFHADHPDVEITLGT-ATSDVLIAELGDGRLD
MSMEG_3840|M.smegmatis_MC2_155 VVGTVTAHDFD-MAGLLAEFHGAHQLVDITLGT-DESDALIDGLLAGRLD
MAB_2238|M.abscessus_ATCC_1997 AFGTVTSHGAD-LSSLLSAFHRDHPAISITLTE-AGCEELVAGLREGRLD
MMAR_4157|M.marinum_M ALGFLHSLGMQTVPHLIRLHHSRFPQARFELHQ-RSGQGVLSDLVSGVTD
MUL_4020|M.ulcerans_Agy99 ALGFLHSLGMQTVPHLIRLHHSRFPQARFELHQ-RSGQGVLSDLVSGVTD
MLBr_02041|M.leprae_Br4923 RLGLIPTVAPYVLPTMLTGLTHRLPTLTLRVVE-DQTEHLLTALREGALD
MAV_2838|M.avium_104 RLGLIPTVAPYVLPTVLAGIAERRPGLTLRVTE-DQTERLLAVLREGALD
Mb0121|M.bovis_AF2122/97 RLGTVPTASTT-ASLVLSAFCSAHPLAKVQVCSRLAATELYRRLREFELD
Rv0117|M.tuberculosis_H37Rv RLGTVPTASTT-ASLVLSAFCSAHPLAKVQVCSRLAATELYRRLREFELD
TH_3504|M.thermoresistible__bu VIGHTPALSGAEAYALIEPAVTAYPSTSFTLRQ-LYPDALTDAVFDGTVQ
.* :: . : : : :
Mvan_3362|M.vanbaalenii_PYR-1 AAIVSIGADEDPAGLEYEVITDEALEAAVTAQHRLARRKSLSLKQLC--E
Mflv_3565|M.gilvum_PYR-GCK LAIVSIAVDEDLPGLEVAVITDEAVEAAVADGHPLAHRRSLSLEQLS--R
MSMEG_3840|M.smegmatis_MC2_155 AAIISVGSG-LPEGLAAEVVTDQRIVAAVGDGHPWRRRRRIALTDLA--G
MAB_2238|M.abscessus_ATCC_1997 GAIVALGAQ-RLAGLGLQVIDDQPLVAAVAADDLLATRRTITLQRLC--E
MMAR_4157|M.marinum_M VSLSFPAAFEGYGEIAWQTLFTQQLYAVVSTDHPLAGRRLIRFPELA--A
MUL_4020|M.ulcerans_Agy99 VSLSFPAAFEGYGEIAWQTLFTQQLYAVVSTDHPLAGRRLIRFPELA--A
MLBr_02041|M.leprae_Br4923 AAMIALPVE--TAGIAEIPIYDEDFVLALPPGHPLSGKRRVPTTALA--Q
MAV_2838|M.avium_104 AALIALPAE--TAGVTAIPIYDEDFVLALPPGHPLAGKRRVPATALA--D
Mb0121|M.bovis_AF2122/97 AVIVHPETQ-DSDDVDLVPLYEEQYVLLSPADMLPPGTSTLVWRDAA--Q
Rv0117|M.tuberculosis_H37Rv AVIVHPETQ-DSDDVDLVPLYEEQYVLLSPADMLPPGTSTLVWRDAA--Q
TH_3504|M.thermoresistible__bu LGLRRGVVP--TPELATAVIGYHRVRVAFVDSHPLAARESVDITELAREQ
: : : . : .
Mvan_3362|M.vanbaalenii_PYR-1 YPVISLPAGTGLRSRLDDACAAAGLRPRIA-FEATSPLELADLARHGLGV
Mflv_3565|M.gilvum_PYR-GCK HPLISLPAGTGLRTRLDTACAAAGLRPRIA-FEATSPAELAELARHGLGV
MSMEG_3840|M.smegmatis_MC2_155 VPLITLPRGTGIRRQLDRACRAAGVDVRVA-FEATTPGALADLAERGLGA
MAB_2238|M.abscessus_ATCC_1997 HTLICLPTGSGVRSILESECRSLGLSPNVA-FEASSPRALVSLVEQGLGI
MMAR_4157|M.marinum_M YPFVVLDRDHTLRRIVDAACARHGIDPTLA-FEGTDVGTLRGLIGANLGV
MUL_4020|M.ulcerans_Agy99 YPFVVLDRDHTLRRIVDAACARHGIDPTPA-FEGTDVGTLRGLIGANLGV
MLBr_02041|M.leprae_Br4923 LPLLLLDEGHCLRDQTLDICRKSGVQAELANTRAASLATAVQCVTGGLGV
MAV_2838|M.avium_104 LPLLLLDEGHCLRDQALDVCHKAGVRAELANTRAASLATAVQCVTGGLGV
Mb0121|M.bovis_AF2122/97 LPLALLTADMRDRQVIDAAFADHAVSAIPQ-VETDSVASLFAQVATGNWA
Rv0117|M.tuberculosis_H37Rv LPLALLTADMRDRQVIDAAFADHAVSAIPQ-VETDSVASLFAQVATGNWA
TH_3504|M.thermoresistible__bu VALWAPPGTSYYSDFLMGACRRAGFEPDYVVSRVQGAATVAAPLTTG-GI
.. .. . .
Mvan_3362|M.vanbaalenii_PYR-1 AILPRSMARGSAG--------LHPLRLSPELRGRLVWAWRTDISG-PAAR
Mflv_3565|M.gilvum_PYR-GCK AILPQSMARGGSG--------LHALRLAPELRGRLVWAWRADGAN-PAAR
MSMEG_3840|M.smegmatis_MC2_155 AVLPESVARARAA--------LHPLPITPELRGRLVFAWRATGPMSPAAR
MAB_2238|M.abscessus_ATCC_1997 AVVPAPYVRNRAG--------VLALDIRPALRGRLALAWRLDGPVGPAAR
MMAR_4157|M.marinum_M GVLPQATARPADI--------IEIAIDDAQLVRPIAIGWIADRCLPSSAV
MUL_4020|M.ulcerans_Agy99 GVLPQATARPADI--------IEIAIDDAQLVRPIAIGWIADRCLPSSAV
MLBr_02041|M.leprae_Br4923 TLLPQSAAPVESVR----SKLGLAQFAAPCPGRRIGLAFRSASGRSASYR
MAV_2838|M.avium_104 TLIPQSAVPVEASR----SRLGLAQFAAPRPGRRIGLVFRSSSGRDDSYR
Mb0121|M.bovis_AF2122/97 SIVPHTWLWAMPMSGPTGGEIRAVELVDPVLKAQIALATNALGPGSPVAR
Rv0117|M.tuberculosis_H37Rv SIVPHTWLWAMPMSGPTGGEIRAVELVDPVLKAQIALATNALGPGSPVAR
TH_3504|M.thermoresistible__bu AFVTAAAGPTMNGR-------VQVVDLTPPLLVPVQALWQRHTLSAIRDR
.:. . . :
Mvan_3362|M.vanbaalenii_PYR-1 LLNARARAMIAATG--TPAVIPGA--
Mflv_3565|M.gilvum_PYR-GCK VLGERARAMAVSPGSGAPAVVPGA--
MSMEG_3840|M.smegmatis_MC2_155 ALVTMARRRLGLGD---PDRVGGHE-
MAB_2238|M.abscessus_ATCC_1997 AFLERARAVL----------------
MMAR_4157|M.marinum_M AFRETALGCYGHRVSAAGV-------
MUL_4020|M.ulcerans_Agy99 AFRETAFGCYGHRDSAAGV-------
MLBr_02041|M.leprae_Br4923 QIAKIIGELISTEHHVRLVA------
MAV_2838|M.avium_104 ELAGLIGELISSQHQVRLVK------
Mb0121|M.bovis_AF2122/97 ALITCAQALALNEFFDTQLRGITRRR
Rv0117|M.tuberculosis_H37Rv ALITCAQALALNEFFDTQLRGITRRR
TH_3504|M.thermoresistible__bu ILEGLG--------------------
: