For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NSRTPSSRSGRSRSREKDTLSREERKEATRRAIVAAALSLLAERSFSGLSLREVTREAGIVPAAFYRHFE SMEALGLVLIDESFRALREMLRGARAGKLDPNRVIESSVDILIDSVQQRREHWRFIGRERSTGVTVLRYA IRTEIRLITSELAIDLARFPNLNTWSSEDLNILASVFVNSMIVIAESLEDATDPASIAEIRRIAIKQLRM ITVGVAGWRSS*
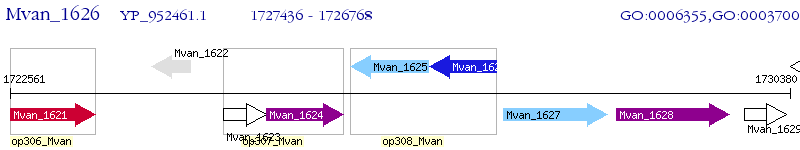
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1626 | - | - | 100% (222) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_4249 | - | 1e-08 | 31.96% (97) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_4778 | - | 2e-08 | 28.57% (147) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1047 | - | 1e-08 | 25.15% (167) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4816 | - | 1e-104 | 88.99% (227) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1019 | - | 1e-08 | 25.15% (167) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_01070 | - | 8e-07 | 37.35% (83) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_1424c | - | 8e-35 | 40.49% (205) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0334 | - | 8e-93 | 78.28% (221) | TetR family transcriptional regulator |
| M. avium 104 | MAV_5169 | - | 6e-86 | 74.65% (213) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_1741 | - | 2e-90 | 75.78% (223) | TetR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_0912 | - | 1e-08 | 25.15% (167) | transcriptional regulator, TetR family protein |
| M. ulcerans Agy99 | MUL_4787 | - | 4e-93 | 78.28% (221) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1047|M.bovis_AF2122/97 ------------------------------------------MTGTER--
Rv1019|M.tuberculosis_H37Rv ------------------------------------------MTGTER--
TH_0912|M.thermoresistible__bu ------------------------------------------MTGSER--
Mvan_1626|M.vanbaalenii_PYR-1 ----------------MNSRTPSSRSGRS-RSREKD-----TLSREERKE
Mflv_4816|M.gilvum_PYR-GCK MGSRQEISAACGRFHTVNSRTPSSRRGHSSRSRSKDGNSRETLSREERKE
MMAR_0334|M.marinum_M ----------------MNERTPSSRPHRS--GRSRD-----SPSREERKE
MUL_4787|M.ulcerans_Agy99 ----------------MNERTPSSRPHRS--SRSRD-----SPSREERKE
MAV_5169|M.avium_104 -----------------------MRGSGR--ERSRE-----SQSREERKE
MSMEG_1741|M.smegmatis_MC2_155 ----------------MNRRTPSTRTRRSETSRSRD-----TLTREERKE
MAB_1424c|M.abscessus_ATCC_199 -----------------------MR-----------------HSRAEKKL
MLBr_01070|M.leprae_Br4923 ----------------MIVVGNFTQSNYVPASRGRR-------SSQLSGD
:
Mb1047|M.bovis_AF2122/97 --RHQLIGIARSLFAERGYDGTSIEEIAQRANVSKPVVYEHFGGKEGLYA
Rv1019|M.tuberculosis_H37Rv --RHQLIGIARSLFAERGYDGTSIEEIAQRANVSKPVVYEHFGGKEGLYA
TH_0912|M.thermoresistible__bu --RQQLIEIAKSLFAERGYEGTSIEEIALRANVSKPVVYEHFGGKEGLYA
Mvan_1626|M.vanbaalenii_PYR-1 ATRRAIVAAALSLLAERSFSGLSLREVTREAGIVPAAFYRHFESMEALGL
Mflv_4816|M.gilvum_PYR-GCK ATRRAIVDAALHLLAERGFSALSLREVTREAGIVPAAFYRHFDSMEALGL
MMAR_0334|M.marinum_M ATRRAIIAAALKLVHDRSFSALSLREVTREAGIVPAAFYRHFESMEALGL
MUL_4787|M.ulcerans_Agy99 ATRRAIIAAALKLVHDRSFSALSLREVTREAGIVPAAFYRHFESMEALGL
MAV_5169|M.avium_104 ATRRAIIAAALKLLQDRSFSSLSLREVTREVGIVPAAFYRHFESMESLGL
MSMEG_1741|M.smegmatis_MC2_155 ATRRALIAAALKLLKDHSFSALSLREVTREAGIVPAAFYRHFDSMDALGL
MAB_1424c|M.abscessus_ATCC_199 HTRRTLLDGTLELLKVRGFAAVSLREVAKSAGLVPTAFYRHFESMEDLGT
MLBr_01070|M.leprae_Br4923 DREQAILAVAERLLAERPLGDFSVDELAKGAGISRPTFYFYFPSKNAVLL
.: :: : *. : *: *:: ..: ...* :* . : :
Mb1047|M.bovis_AF2122/97 VVVDREMSALLDGITSSL---TNNRSRVRVERVALALLTYVEERTDGFRI
Rv1019|M.tuberculosis_H37Rv VVVDREMSALLDGITSSL---TNNRSRVRVERVALALLTYVEERTDGFRI
TH_0912|M.thermoresistible__bu VVVDREMSALLDGITSSL---TNNRSRVRVERVALALLTYVEERTDGFRI
Mvan_1626|M.vanbaalenii_PYR-1 VLIDESFRALREMLRGAR--AGKLDPNRVIESSVDILIDSVQQRREHWRF
Mflv_4816|M.gilvum_PYR-GCK VLIDESFRSLREMLRGAR--AGKLDPNRVIESSVDILIDGVQQRREHWRF
MMAR_0334|M.marinum_M VLIDESFRTLRDTLRGAR--AGRLDPNRVIESSVEILIGSVAERREHWRF
MUL_4787|M.ulcerans_Agy99 VLIDESFRTLRDTLRGAR--AGRLDPNRVIESSVEILIGSVAERREHWRF
MAV_5169|M.avium_104 VLIDESFRSLRDTLRDAR--AGKLDPNRVIESSVEILVASVADRREHWRL
MSMEG_1741|M.smegmatis_MC2_155 VLIDESFRSLRDMLRGAR--AGKLDPHHVIESSAEILVSSFNERREHWRF
MAB_1424c|M.abscessus_ATCC_199 VLAEESVHALRDTFRQARREAGSDNPAKILR----LMVDRVNEHANNFRF
MLBr_01070|M.leprae_Br4923 SLLDDLNMKSRSAVEALA-EQLPADPAAVWRSAITAFFEVCGTHRAVAVA
: : . . . . :. :
Mb1047|M.bovis_AF2122/97 MIRDSPASISSG-TYSSLLNDAVSQVSSILAGDFARR----GLDPDLAPL
Rv1019|M.tuberculosis_H37Rv MIRDSPASISSG-TYSSLLNDAVSQVSSILAGDFARR----GLDPDLAPL
TH_0912|M.thermoresistible__bu LIRDSPAAITSG-TYSTLLNDAVNQVSSILAGDFSRR----GLDPEMAPL
Mvan_1626|M.vanbaalenii_PYR-1 IGRERSTGVTV---LRYAIRTEIRLITSELAIDLARFPNLNTWSSEDLNI
Mflv_4816|M.gilvum_PYR-GCK IGRERSTGVTV---LRYAIRTEIRLITSELAIDLARFPALNTWSSEDLNI
MMAR_0334|M.marinum_M IVRERSTGLSV---LRYAIRTEIRLITSELATDLARFPGLNEWSTEDLNI
MUL_4787|M.ulcerans_Agy99 IVRERSTGLSV---LRYAIRTDIRLITSELATDLARFPGLNEWSTEDLNI
MAV_5169|M.avium_104 IARERNSGLSV---LRYAIRTEIRLITSELATDLARFPGLHEWTTEDLNV
MSMEG_1741|M.smegmatis_MC2_155 IARERSGGASA---LRYAIRTEIRLITSELAIDLARFPALKDWSSEDLNI
MAB_1424c|M.abscessus_ATCC_199 LVRERHGGSPQ---IRRAINNEMRMLTNDVVVELARNPATNHHTTEDLEI
MLBr_01070|M.leprae_Br4923 GAAAKATSPEVRRLWSTVMQQWIDYNTAAIQAERKCGVAPDTIPAEDLAV
. :. : : : : .: :
Mb1047|M.bovis_AF2122/97 YAQALVGSVSMTAQWWLDAREP------KKEVVAAHLVNLVWNGLTHLEA
Rv1019|M.tuberculosis_H37Rv YAQALVGSVSMTAQWWLDAREP------KKEVVAAHLVNLVWNGLTHLEA
TH_0912|M.thermoresistible__bu YAQALVGSVSMTAQWWLDVREP------KKEVVAAHLVNLCWNGLTHLEP
Mvan_1626|M.vanbaalenii_PYR-1 LASVFVNSMIVIAESLEDATDP--ASIAEIRRIAIKQLRMITVGVAGWRS
Mflv_4816|M.gilvum_PYR-GCK LASVFVNSMIAIAEQLEDVTDT--ASVDEIRRTAVKQLRMITVGVAGWRS
MMAR_0334|M.marinum_M LATLFVNAMIVTAEAIEDAQSS--EALEEIRRIAVKQLRMVAIGVAGWKS
MUL_4787|M.ulcerans_Agy99 LATMFVNAMIVTAEAIEDAQSS--EALEEIRRIAVKQLRMVAIGVAGWKS
MAV_5169|M.avium_104 LATLFVNAMIVIAEAIEDSQSA--EAREDIRRLAVKQLRMIATGIAGWRS
MSMEG_1741|M.smegmatis_MC2_155 LANLFVNSMISLAEALEDAADA--ESIEAIHRTAVKQMRMIAVGVNGWRS
MAB_1424c|M.abscessus_ATCC_199 AAELIASSILNTIGMLVDTDHPTKADADAALNRALRQLDLVTSGLAHTRT
MLBr_01070|M.leprae_Br4923 ALQLMTERVMAATFSDEKPLIP-------DKKVIDTLVHIWLASIYGS--
:. : . . : : .:
Mb1047|M.bovis_AF2122/97 DPRLQDE---------------
Rv1019|M.tuberculosis_H37Rv DPRLQDE---------------
TH_0912|M.thermoresistible__bu DPVLHEE---------------
Mvan_1626|M.vanbaalenii_PYR-1 S---------------------
Mflv_4816|M.gilvum_PYR-GCK G---------------------
MMAR_0334|M.marinum_M RP--------------------
MUL_4787|M.ulcerans_Agy99 RP--------------------
MAV_5169|M.avium_104 TP--------------------
MSMEG_1741|M.smegmatis_MC2_155 TD--------------------
MAB_1424c|M.abscessus_ATCC_199 MTTIIAEPAARPSESSATTAAP
MLBr_01070|M.leprae_Br4923 ----------------------