For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLAGSWIDGAPVNTRGPVHQVINPATGAPVAEYALAQPADVDAAVASARAALGGWATATPAERSAVLAKL AKLADEATADLVAEEVSQTGKPVRLATEFDVPGSVDNIDFFAGAARHLEGKASGEYSADHTSSIRREAVG VVATITPWNYPLQMAVWKVLPALAAGCSVVIKPAEITPLTTLTLARLAAEAGLPAGVFNVVTGSGADVGT ALAGHADVDMVTFTGSTPVGRKVMLAAAAHGHRTQLELGGKAPFVVFDDADLDAAIQGAVAGALINTGQD CTAATRAIVARDLYDDFVAGVAEVMSNIVVGDPGDPDTDLGPLITLAHRAKVAGMVERAPGEGGRIVTGG VAPDGPGSFYRPTLIADVSEQSEVWRDEIFGPVLAVRSFTDDDDALRQANDTAYGLAASAWTRDVYRAQR ASREIHAGCVWINDHIPIISEMPHGGFGASGFGKDMSQYSLEEYLSIKHVMSDITGVADKTWHRTIFTKR
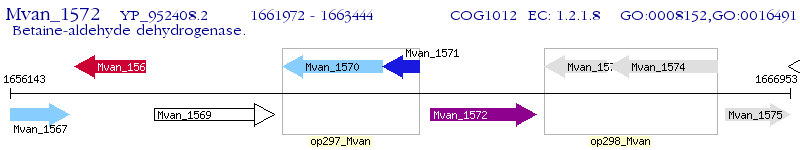
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1572 | - | - | 100% (490) | gamma-aminobutyraldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_2580 | - | e-111 | 44.49% (472) | gamma-aminobutyraldehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0014 | - | 3e-93 | 41.13% (479) | betaine-aldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2883c | aldC | 5e-89 | 41.02% (451) | aldehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4860 | - | 0.0 | 90.80% (489) | gamma-aminobutyraldehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv2858c | aldC | 5e-89 | 41.02% (451) | aldehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 3e-70 | 37.25% (459) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3328 | - | 0.0 | 81.89% (486) | gamma-aminobutyraldehyde dehydrogenase |
| M. marinum M | MMAR_3556 | - | 0.0 | 78.12% (489) | aldehyde dehydrogenase NAD dependent |
| M. avium 104 | MAV_2101 | - | 2e-95 | 41.96% (460) | aldehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1665 | - | 0.0 | 85.69% (489) | gamma-aminobutyraldehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_2957 | - | 2e-86 | 40.75% (454) | PUTATIVE aldehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_2789 | - | 0.0 | 76.69% (489) | gamma-aminobutyraldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2883c|M.bovis_AF2122/97 -------------------------MSTTQLINPATEEVLASVDHTDANA
Rv2858c|M.tuberculosis_H37Rv -------------------------MSTTQLINPATEEVLASVDHTDANA
Mvan_1572|M.vanbaalenii_PYR-1 -----MLAGSWID-----GAPVNTRGPVHQVINPATGAPVAEYALAQPAD
Mflv_4860|M.gilvum_PYR-GCK ----MTVASSWID-----GAPVKTGGAQHTVINPATGESVAEFALAQPGD
MSMEG_1665|M.smegmatis_MC2_155 --MAVKVASSWIN-----GASVETSGETHQIIDPATGKSVSEYNLATAAD
MAB_3328|M.abscessus_ATCC_1997 ----MSVPSSWIN-----GRPVATHGDTHRVINPATGEAVADLKLATTAD
MMAR_3556|M.marinum_M MTTTVSVAGSWIN-----GAAHSTGGRQHAVINPATQETIAELALATPAD
MUL_2789|M.ulcerans_Agy99 MTTTVSVAGSWIN-----GAAHSTGGRQHAVINPATQETIAELALATPAD
MAV_2101|M.avium_104 --MSTTIP----------------------VFNPSTEEQIAEVPDSDQAA
TH_2957|M.thermoresistible__bu --MTSTLERPEVTLRIGGEKRRTASGGTFDHINPCTGAPDAAIPLAGPAE
MLBr_02573|M.leprae_Br4923 --------------------------MSIATINPATGETVKTFTPTTQDE
::*.* :
Mb2883c|M.bovis_AF2122/97 VDDAVQRARAAQRR--WARLAPAQRAAGLRAFAAAVQAHLDELAALEVAN
Rv2858c|M.tuberculosis_H37Rv VDDAVQRARAAQRR--WARLAPAQRAAGLRAFAAAVQAHLDELAALEVAN
Mvan_1572|M.vanbaalenii_PYR-1 VDAAVASARAALGG--WATATPAERSAVLAKLAKLADEATADLVAEEVSQ
Mflv_4860|M.gilvum_PYR-GCK VDRAVVSARAALPG--WKGATPAERSAVLAKLAKLADDATADLVAEEVSQ
MSMEG_1665|M.smegmatis_MC2_155 VDTAVAAARAAFPA--WAGATPAERSAVLAKLAKLADEHAEQFVAEEVSQ
MAB_3328|M.abscessus_ATCC_1997 VDAAVAAARAAGPA--WASSTPVDRSSVLAKLAAVLSANADTLVADEVAQ
MMAR_3556|M.marinum_M VDRAVVAARRALPQ--WAGVTPAERSAVLAKLADLLEQRAPELIAEEVCQ
MUL_2789|M.ulcerans_Agy99 VDHAVVAARRALPQ--WAGVTTAERSAVLAELADLLEQRAPELIAEEVCQ
MAV_2101|M.avium_104 VDAAVARARETFESGVWRKAPASHRANVLFRAARIIEERKDELAALESQD
TH_2957|M.thermoresistible__bu IDDAVKAAHDAFHS--WRQTNPAERRRLLFRLAELIEQHTDDFVRLGTFD
MLBr_02573|M.leprae_Br4923 VEDAIARAYARFDN--YRHTTFTQRAKWANATADLLEAEANQSAALMTLE
:: *: * : .* * . :
Mb2883c|M.bovis_AF2122/97 SGHPI-VSAEWEAGHVRDVLAFYAASPERLSGRQIP---------VAGGV
Rv2858c|M.tuberculosis_H37Rv SGHPI-VSAEWEAGHVRDVLAFYAASPERLSGRQIP---------VAGGV
Mvan_1572|M.vanbaalenii_PYR-1 TGKPVRLATEFDVPGSVDNIDFFAGAARHLEGKAS-------GEYSADHT
Mflv_4860|M.gilvum_PYR-GCK TGKPVRLATEFDVPGSVDNIDFFAGAARHLEGKAS-------AEYSGDHT
MSMEG_1665|M.smegmatis_MC2_155 TGKPVRLATEFDVPGSVDNIDFFAGAARHLEGKAT-------AEYSGDHT
MAB_3328|M.abscessus_ATCC_1997 TGKPVRLASEFDVPGSIDNVEFFAGAARHLEGKAT-------AEYSADHT
MMAR_3556|M.marinum_M TGKPLRLAEEFDVPGSVDNIRFFAGAARRLPGQAT-------AEYSADHT
MUL_2789|M.ulcerans_Agy99 TGKPLRLAEEFDVPGSVDNIRFFAGAARRLPGQAT-------AEYSADHT
MAV_2101|M.avium_104 NGMNLTAAGHI-IPVSVDMLKYYAGWVGKIHGESANLVSDGLLGTWETYH
TH_2957|M.thermoresistible__bu NGTPVGLVAGL-VAMSAEWTRYYAGWADKITSEVS-----GTYGTTGEFS
MLBr_02573|M.leprae_Br4923 MGKTLQSAKDEAMKCAKG-FRYYAEKAETLLADEP-----AEAAAVGASK
* : ::* : .
Mb2883c|M.bovis_AF2122/97 DVTFNEPMGVVGVITPWNFPMVIASWAIAPALAAGNAVLVKPAELTPLTT
Rv2858c|M.tuberculosis_H37Rv DVTFNEPMGVVGVITPWNFPMVIASWAIAPALAAGNAVLVKPAELTPLTT
Mvan_1572|M.vanbaalenii_PYR-1 SSIRREAVGVVATITPWNYPLQMAVWKVLPALAAGCSVVIKPAEITPLTT
Mflv_4860|M.gilvum_PYR-GCK SSIRREAVGVVATITPWNYPLQMAVWKVIPALAAGCSVVIKPAEITPLTT
MSMEG_1665|M.smegmatis_MC2_155 SSIRREAVGVVATITPWNYPLQMAVWKVIPALAAGCAVVIKPAEITPLTT
MAB_3328|M.abscessus_ATCC_1997 SSIRREAAGVVATITPWNYPLQMAVWKVLPALAAGCTVVIKPSELTPLTT
MMAR_3556|M.marinum_M SSIRREAIGVVATITPWNYPLQMAVWKVMPALAAGCTVVIKPSELTPLTT
MUL_2789|M.ulcerans_Agy99 SSIRREAIGVVATITPWNYPLQMAVWKVMPALAAGCTVVIKPSELTPLTT
MAV_2101|M.avium_104 TFTQLEPVGVVGLIIPWNGPFFVAMLKVAPALAAGCSAVLKPAEETPLTA
TH_2957|M.thermoresistible__bu -YTLSQPYGVIGVIITWNGPLISLAMKVPAALAAGNTVVVKPSELTPFSA
MLBr_02573|M.leprae_Br4923 ALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASNVPQCA
:. **: : .** *: .** ** ::* :. .* :
Mb2883c|M.bovis_AF2122/97 MRLGELAVEAGLDEDLLQVLPGKGTVVGERFVTHPDIRKIVFTGSTEVGK
Rv2858c|M.tuberculosis_H37Rv MRLGELAVEAGLDEDLLQVLPGKGTVVGERFVTHPDIRKIVFTGSTEVGK
Mvan_1572|M.vanbaalenii_PYR-1 LTLARLAAEAGLPAGVFNVVTGSGADVGTALAGHADVDMVTFTGSTPVGR
Mflv_4860|M.gilvum_PYR-GCK LTLARLASEAGLPDGVLNVVTGSGADVGAALAGHADVDMVTFTGSTPVGR
MSMEG_1665|M.smegmatis_MC2_155 LTLARLASEAGLPDGVFNVVTGSGAEVGTALAGHRDVDVVTFTGSTAVGR
MAB_3328|M.abscessus_ATCC_1997 LTLARLAAEAGLPEGVLNVVTGRGDDVGYALASHPDVDLVTFTGSTAVGR
MMAR_3556|M.marinum_M LTLARLAAEAGLPDGVLNVVTGLGADVGAALAAHPGVDLVTFTGSTAVGR
MUL_2789|M.ulcerans_Agy99 LTLARLAAEAGLPDGVLNVVTGLGADVGAALAAHPGVDLVTFTGSTAVGR
MAV_2101|M.avium_104 LKLEEIFREAGLPDGVLNVITGYGETTGAALTAHPDVDKISFTGSTEVGR
TH_2957|M.thermoresistible__bu TLFADLAAEAGIPDGVINVVPGTGD-AGAALVAHPLVKKVTFTGGPDTAR
MLBr_02573|M.leprae_Br4923 LYLADVIARGGFPEDCFQTLLISANGVEAILR-DPRVAAATLTGSEPAGQ
: : ..*: . ::.: . . : . : :**. ..:
Mb2883c|M.bovis_AF2122/97 RVMAGAAAQVKRVTLELGGKSANIVFHDCDLERAA-TTAPAGVFDNAGQD
Rv2858c|M.tuberculosis_H37Rv RVMAGAAAQVKRVTLELGGKSANIVFHDCDLERAA-TTAPAGVFDNAGQD
Mvan_1572|M.vanbaalenii_PYR-1 KVMLAAAAHGHRTQLELGGKAPFVVFDDADLDAAI-QGAVAGALINTGQD
Mflv_4860|M.gilvum_PYR-GCK RVMLAAAAHGHRTQLELGGKAPFVVFDDADLDAAI-QGAVAGALINTGQD
MSMEG_1665|M.smegmatis_MC2_155 KVMASAAVHGHRTQLELGGKAPFVVFSDADLDAAV-QGAVAGALINTGQD
MAB_3328|M.abscessus_ATCC_1997 KVMAAAAVHGHRTQLELGGKAPFVVFDDADMDAAI-AGAVAGAIINSGQD
MMAR_3556|M.marinum_M KVMAAAATHGRRTQLELGGKAPFVVFDDADLDAAI-QGAVAGALINSGQD
MUL_2789|M.ulcerans_Agy99 KVMAAAATHGRRTQLELGGKAPFVVFDDADLDAAI-QGAVAGAVINSGQD
MAV_2101|M.avium_104 LIVKAAAGNLKRLTLELGGKSPLIMFDDANLDKAI-MGAGMGLLAGSGQN
TH_2957|M.thermoresistible__bu KILQSCAEHIKPSVMELGGKSGNIIFEDANLDTACSVGTLMSVGMMSGQG
MLBr_02573|M.leprae_Br4923 AVGAIAGNEIKPTVLELGGSDPFIVMPSADLDAAV-ATAVTGRVQNNGQS
: .. . : :****. ::: ..::: * : . **.
Mb2883c|M.bovis_AF2122/97 CCARSRILVQRSVYDRFMELLEPAVHSIVVGDPGSRATEMGPLVSRAHRD
Rv2858c|M.tuberculosis_H37Rv CCARSRILVQRSVYDRFMELLEPAVHSIVVGDPGSRATEMGPLVSRAHRD
Mvan_1572|M.vanbaalenii_PYR-1 CTAATRAIVARDLYDDFVAGVAEVMSNIVVGDPGDPDTDLGPLITLAHRA
Mflv_4860|M.gilvum_PYR-GCK CTAATRAIVAEGLYDDFVAGVAEVMSKVVIGDPADPDTDLGPLISAVHRE
MSMEG_1665|M.smegmatis_MC2_155 CTAATRAIVARELYDDFVAGVAEVMGKIVVGDPHDPDTDLGPLISYAHRD
MAB_3328|M.abscessus_ATCC_1997 CTAATRALVARDLYDDFVAGVGEVMSKVVVGDPLDPDTDIGPLISAAHRA
MMAR_3556|M.marinum_M CTAATRAIVARELYHDFVAGVADVMSTVVIGDPMDPDTDIGPLISLAHRD
MUL_2789|M.ulcerans_Agy99 CTAATRAIVARELCHDFVAGVADVMSTVVIGDPMDPDTDIGPLISLAHRD
MAV_2101|M.avium_104 CSCTSRIYVQRGIYDQVVEGLAEFAMMLPMGGSDDPNSVLGPLISEKQRQ
TH_2957|M.thermoresistible__bu CAFPTRMIVARPLYDEVLTRVSALAPMVKVGDPLEPDTVAGPVVNEAALQ
MLBr_02573|M.leprae_Br4923 CIAAKRFIAHADIYDEFVEKFVARMETLRVGDPTDPDTDVGPLATESGRA
* .* . : . .: . : :*.. . : **: .
Mb2883c|M.bovis_AF2122/97 KVAGYVP----DDAPVAFRGTAPAG---RGFWFPPTVLTPKRGD-RTVTD
Rv2858c|M.tuberculosis_H37Rv KVAGYVP----DDAPVAFRGTAPAG---RGFWFPPTVLTPKRGD-RTVTD
Mvan_1572|M.vanbaalenii_PYR-1 KVAGMVERAPGEGGRIVTGGVAPD---GPGSFYRPTLIADVSEQSEVWRD
Mflv_4860|M.gilvum_PYR-GCK KVAGMVERAPGEGGRVVTGGVAPD---GPGSFYRPTLIADVAESSEVWRD
MSMEG_1665|M.smegmatis_MC2_155 KVAGMVARAPQQGGRVVTGGEAPD---LPGAFYRPTLIADVDESSEVYRD
MAB_3328|M.abscessus_ATCC_1997 KVSSIVDRAPAQGGRIVTGATAPD---LPGSFYRPTLIADVAETSEVYRD
MMAR_3556|M.marinum_M RVADIVAKAPAQGARIVVGGRIPD---LAGAFYLPTLLADVGEQSQAYRE
MUL_2789|M.ulcerans_Agy99 RVADILAKAPAQGARIVVGGRIPD---LAGAFYLPTLLADVGEQSQAYRE
MAV_2101|M.avium_104 RVEGIVNDGVAAGAKVITGGKPMD---RKGYFYEATIITNTTPDMRLIRE
TH_2957|M.thermoresistible__bu RILGMIERAKQDGARLLTGGERLGGDLADEYFVAPTVFADVDPYSELAQK
MLBr_02573|M.leprae_Br4923 QVEQQVDAAAAAGAVIRCGGKRPD---RPGWFYPPTVITDITKDMALYTD
:: : .. : . : .*::: .
Mb2883c|M.bovis_AF2122/97 EIFGPVVVVLTFDDEADAISLANDTAYGLSGSIWTDDLSRALRVARAVES
Rv2858c|M.tuberculosis_H37Rv EIFGPVVVVLTFDDEADAISLANDTAYGLSGSIWTDDLSRALRVARAVES
Mvan_1572|M.vanbaalenii_PYR-1 EIFGPVLAVRSFTDDDDALRQANDTAYGLAASAWTRDVYRAQRASREIHA
Mflv_4860|M.gilvum_PYR-GCK EIFGPVLTVRSFTDDDDAIRQANDTKYGLAASAWTRDVYRAQRASREIHA
MSMEG_1665|M.smegmatis_MC2_155 EIFGPVLTVRSFTDDDDALRQANDTEYGLAASAWTRDVYRAQRASREINA
MAB_3328|M.abscessus_ATCC_1997 EIFGPVLTVRPFTDDDDALRQANDTAYGLAASAWTRDVYRAQRASREIRA
MMAR_3556|M.marinum_M EIFGPVLTVRAHDGDDDALRQANDTDYGLAASAWTRDVYRAQRASREISA
MUL_2789|M.ulcerans_Agy99 EIFGPVLTVRAHDGDDDALRQANDTDYGLAASAWTRDVYRAQRASREISA
MAV_2101|M.avium_104 EIFGPVGCVIPFDDEDEVIAAANDTHYGLAGAIWTENLSRAHRVVNELRA
TH_2957|M.thermoresistible__bu EVFGPVLSIIPFDTEEQAVEIANSTPYGLSGYVFTNDLKRAHRVAEQLET
MLBr_02573|M.leprae_Br4923 EVFGPVASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQRHFIDNIVA
*:**** : :.: ** * :**.. :* : . : : :
Mb2883c|M.bovis_AF2122/97 GNLSVNSHSSVRFNTPFGGFKQSGVGRELGPDAPLQFTETKNVFIAVGEE
Rv2858c|M.tuberculosis_H37Rv GNLSVNSHSSVRFNTPFGGFKQSGVGRELGPDAPLQFTETKNVFIAVGEE
Mvan_1572|M.vanbaalenii_PYR-1 GCVWINDHIPIISEMPHGGFGASGFGKDMSQYSLEEYLSIKHVMSDITGV
Mflv_4860|M.gilvum_PYR-GCK GCVWINDHIPIISEMPHGGFGASGFGKDMSQYSLEEYLSIKHVMSDITSA
MSMEG_1665|M.smegmatis_MC2_155 GCVWINDHIPIISEMPHGGVGASGFGKDMSDYSFEEYLTIKHVMSDITGV
MAB_3328|M.abscessus_ATCC_1997 GCVWINDHIPIISEMPHGGFGASGFGKDMSDYSLEEYLTVKHVMSDITGT
MMAR_3556|M.marinum_M GCVWINDHIPIVSEMPHGGVAASGFGKDMSEYSFEEYLTIKHVMSDITGV
MUL_2789|M.ulcerans_Agy99 GCVWINDHIPIVSEMPHGGVAASGFGNDMSEYSFDEYLTIKHVMSDITGV
MAV_2101|M.avium_104 GQIWVNSALAADPSMPISGHKQSGWGGERGRKGIEAYFNTKAVYIGL---
TH_2957|M.thermoresistible__bu GEVLINGAAPLTVHRPFGGFGISGIGKEGGREGLAEFLRTKSVSIA----
MLBr_02573|M.leprae_Br4923 GQVFINGMTVSYPELPFGGIKRSGYGRELSTHGIREFCNIKTVWIA----
* : :*. * .* ** * : . . : * *
Mb2883c|M.bovis_AF2122/97 M-------------
Rv2858c|M.tuberculosis_H37Rv M-------------
Mvan_1572|M.vanbaalenii_PYR-1 ADKTWHRTIFTKR-
Mflv_4860|M.gilvum_PYR-GCK VDKDWHRTIFTKR-
MSMEG_1665|M.smegmatis_MC2_155 AEKEWHRTIFNKR-
MAB_3328|M.abscessus_ATCC_1997 ADKDWHRTIFAKR-
MMAR_3556|M.marinum_M AEKPWHRTIFSTRI
MUL_2789|M.ulcerans_Agy99 AEKPWHRTIFSTRI
MAV_2101|M.avium_104 --------------
TH_2957|M.thermoresistible__bu --------------
MLBr_02573|M.leprae_Br4923 --------------