For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDAPGSFRAQQRFSDPMAPVPAEWTAPTPPEGAPPVVTADPAPYLDLSTVALLGNASRGPSAGWRKWLYW GSGRLIDVGESPKVIRRNALVAQVQRPLRDCYRIAMLSQKGGVGKTTITATLGATFATTRGDRVIAVDAN PDRGTLSQKVPLETPATVRHLLRDAEGIAAYSDVRQYTSQGPSRLEVLASESDPAVSEAFSAADYCRAVD VLERFYSVVLTDCGTGMLHSAMSGVIDKADVLVVISSGSVDGARSASATLDWLDAHGHGDMVGNSIAVIN GVRRGARKGAGKGTAKVDLNKVVDHFARRCRAVCQVPFDPHLEEGAEISLDRLRADTRESLLELAAAVAD GFPGARRTDPVG
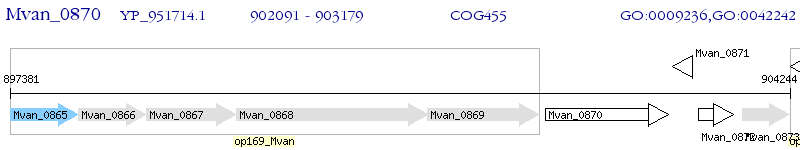
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0870 | - | - | 100% (362) | hypothetical protein Mvan_0870 |
| M. vanbaalenii PYR-1 | Mvan_0080 | - | 9e-58 | 36.22% (370) | hypothetical protein Mvan_0080 |
| M. vanbaalenii PYR-1 | Mvan_0069 | - | 2e-40 | 33.33% (342) | hypothetical protein Mvan_0069 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0543 | - | 1e-125 | 68.84% (337) | hypothetical protein Mb0543 |
| M. gilvum PYR-GCK | Mflv_0048 | - | 1e-168 | 82.27% (361) | hypothetical protein Mflv_0048 |
| M. tuberculosis H37Rv | Rv0530 | - | 1e-125 | 68.84% (337) | hypothetical protein Rv0530 |
| M. leprae Br4923 | MLBr_00798 | - | 8e-47 | 35.01% (337) | hypothetical protein MLBr_00798 |
| M. abscessus ATCC 19977 | MAB_3972c | - | 1e-128 | 64.34% (387) | hypothetical protein MAB_3972c |
| M. marinum M | MMAR_0876 | - | 1e-121 | 65.67% (335) | hypothetical protein MMAR_0876 |
| M. avium 104 | MAV_4615 | - | 1e-124 | 69.94% (336) | hypothetical protein MAV_4615 |
| M. smegmatis MC2 155 | MSMEG_0975 | - | 1e-138 | 65.42% (402) | hypothetical protein MSMEG_0975 |
| M. thermoresistible (build 8) | TH_0884 | - | 1e-126 | 72.84% (313) | conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3237 | - | 8e-06 | 36.43% (129) | Soj family ATPase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0543|M.bovis_AF2122/97 --------------------------------------------------
Rv0530|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0876|M.marinum_M --------------------------------------------------
MAV_4615|M.avium_104 --------------------------------------------------
Mvan_0870|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0048|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0975|M.smegmatis_MC2_155 --------------------------------------------------
TH_0884|M.thermoresistible__bu --------------------------------------------------
MAB_3972c|M.abscessus_ATCC_199 ----------------------------MSDYPAHRAPDGPSPYAADEAQ
MLBr_00798|M.leprae_Br4923 MTTFRERRTMFDAAVASYQSGDKTSAGAAFARLTVENPDMSDSWLGRLAC
MUL_3237|M.ulcerans_Agy99 --------------------------------------------------
Mb0543|M.bovis_AF2122/97 -----------------------------------------MLVTEHPRT
Rv0530|M.tuberculosis_H37Rv -----------------------------------------MLVTEHPRT
MMAR_0876|M.marinum_M -----------------------------------------MDVSDHPSA
MAV_4615|M.avium_104 -------------------------------------------MSEHPTA
Mvan_0870|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0048|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0975|M.smegmatis_MC2_155 --------------------------------------------------
TH_0884|M.thermoresistible__bu --------------------------------------------------
MAB_3972c|M.abscessus_ATCC_199 GGPTAPPWAAQEPAQPVMYGQSTPQQPHEMTQQFQAVPFQDPFAPQQDIA
MLBr_00798|M.leprae_Br4923 GDHNLDTLAGAHQHSEVLYSETRRVGLKDGDLRAEIAAPMYLTLTTWSRA
MUL_3237|M.ulcerans_Agy99 --------------------------------------------------
Mb0543|M.bovis_AF2122/97 GVGAPDS----GNGGTDHPTVQLPP-----------------------VP
Rv0530|M.tuberculosis_H37Rv GVGAPDS----GNGGTDHPTVQLPP-----------------------VP
MMAR_0876|M.marinum_M GVGASNAY--QGEGGKDHPTIQLPP-----------------------VP
MAV_4615|M.avium_104 GVGAPEE-----------QTTQIPP-----------------------AA
Mvan_0870|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0048|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0975|M.smegmatis_MC2_155 -----------MSASSEHPVPDSVD-----------------------VP
TH_0884|M.thermoresistible__bu --------------------------------------------------
MAB_3972c|M.abscessus_ATCC_199 GLGHDPAVPSLNSLGGSFAPLDFTP-----------------------SG
MLBr_00798|M.leprae_Br4923 TIGLAYASALINAGRYDEASVALDDPVVTEDDDAALARQFVMATLFHQTR
MUL_3237|M.ulcerans_Agy99 --------------------------------------------------
Mb0543|M.bovis_AF2122/97 S-------VGAPPAAAGGETPTRSVAGFRTQR------------------
Rv0530|M.tuberculosis_H37Rv S-------VGAPPAAAGGETPTRSVAGFRTQR------------------
MMAR_0876|M.marinum_M HQ---PPQLGEPPVAGGSDAPTRAFAGFRTERRSAARAEGAEAPDVAQWE
MAV_4615|M.avium_104 AAGDDKKDAAEPQPEPGGDAPTKAFAGFRTERRVPEREPAPPTAPRPGGM
Mvan_0870|M.vanbaalenii_PYR-1 ---------------------MDAPGSFRAQQRFSDP------MAPVPAE
Mflv_0048|M.gilvum_PYR-GCK -------------------------MSFRAQQRFSDP------MAPVPAE
MSMEG_0975|M.smegmatis_MC2_155 AP------EHPAAETPEAPSVFAPVPGFRGQQRFSDPA-QAGVSTPLPAE
TH_0884|M.thermoresistible__bu --------------------------------------------------
MAB_3972c|M.abscessus_ATCC_199 SQ------EAGDPTAVGAAGSSGGPGGFRADNRFSDSGEQPAQPTPFTAD
MLBr_00798|M.leprae_Br4923 SWSNVLKVAEVPPQNSNTDDRNELTDAVAALASTAASNLGQFQFALELTE
MUL_3237|M.ulcerans_Agy99 --------------------------------------------------
Mb0543|M.bovis_AF2122/97 -------------LDPTAYGAYYSGPDEGPASP--------------AER
Rv0530|M.tuberculosis_H37Rv -------------LDPTAYGAYYSGPDEGPASP--------------AER
MMAR_0876|M.marinum_M SGPDTGPVAGLPRVDPTAYGAYYNGPPEPPAEP--------------QYR
MAV_4615|M.avium_104 PPWDSTPVTGIPRVDPTAYGAYYAGPDAPPTQI--------------QGP
Mvan_0870|M.vanbaalenii_PYR-1 -WTAPTPPEGAPP------------------------------------V
Mflv_0048|M.gilvum_PYR-GCK -WTAPTPPEGVVP------------------------------------V
MSMEG_0975|M.smegmatis_MC2_155 -WTAPTPPHGIPVSAPQTAQTPPQNSHQPMASGLPPLPP---APLTPPAA
TH_0884|M.thermoresistible__bu --------------------------------------------------
MAB_3972c|M.abscessus_ATCC_199 RWASDTPPSGIPQQAQQPPVGGSRYYTPVGGQ---------------QTG
MLBr_00798|M.leprae_Br4923 RVSTTNPQVAADVALNRAWCLRELGDDNAARAALTAAPVNDIPTTHNTAE
MUL_3237|M.ulcerans_Agy99 --------------------------------------------------
Mb0543|M.bovis_AF2122/97 PPYRLEPVPHTPYPELATTTLLRPVKPPPSEGWRRLLYLLSGRLINAGEG
Rv0530|M.tuberculosis_H37Rv PPYRLEPVPHTPYPELATTTLLRPVKPPPSEGWRRLLYLLSGRLINAGEG
MMAR_0876|M.marinum_M P----ESVPQTPYPELSTSMLLRPVKSPPTEGWRRLVYVLSGQLINLGES
MAV_4615|M.avium_104 PP-RQEHLPHTPYPELSTGMLLRPVQPPPSEGWRLLLYKLSGGLINLGES
Mvan_0870|M.vanbaalenii_PYR-1 VTADP-----APYLDLSTVALLGNASRGPSAGWRKWLYWGSGRLIDVGES
Mflv_0048|M.gilvum_PYR-GCK VAAATPSTELVPYRELSTVALLGALPKAPSAGWRKWVYRASGRLINPGES
MSMEG_0975|M.smegmatis_MC2_155 AAEQPPADFATPYEDLTTTALLGRRKSPPTAGWRRWLYLASFKQVNVGES
TH_0884|M.thermoresistible__bu -------------VDLSTTALLGQRRPAPSRGWRRWLYRATFRTVNLGES
MAB_3972c|M.abscessus_ATCC_199 VGGQIRPELPPSSRDLSTSALLRPHKPAPSAGWRKTVYRASFGTINLGEG
MLBr_00798|M.leprae_Br4923 PASRPQPTTFRHPYDDGRDLLVARRRPAAGDGWRKFVTKATLGRVNPEPS
MUL_3237|M.ulcerans_Agy99 -----MPAGLPAQADAAVRLSCDVHPDVLHNASRSGMNDHPDSGVSVGLT
: . * : . :.
Mb0543|M.bovis_AF2122/97 PRAAHLNDLVAQVNRPLRGCYRIAVLSLKGGVGKTTITATLGATFADLRG
Rv0530|M.tuberculosis_H37Rv PRAAHLNDLVAQVNRPLRGCYRIAVLSLKGGVGKTTITATLGATFADLRG
MMAR_0876|M.marinum_M PRATRYNNLVAQINRPMRGCYRIALVSLKGGVGKTTITATLGSTFASVRG
MAV_4615|M.avium_104 PRAARYNNLVAQVNRPLRGSYRVAFLSLKGGVGKTTIAATLGATFASIRG
Mvan_0870|M.vanbaalenii_PYR-1 PKVIRRNALVAQVQRPLRDCYRIAMLSQKGGVGKTTITATLGATFATTRG
Mflv_0048|M.gilvum_PYR-GCK PKVIRHDDLLARVQQPLQGCYRVAMLSQKGGVGKTTITATLGATFAGARG
MSMEG_0975|M.smegmatis_MC2_155 PKVTHRQSLVSQVSHPLQGCYRIAVLSLKGGVGKTTITATLGSTFASIRG
TH_0884|M.thermoresistible__bu PKVARHNELLNRVRHPLQDCYRIAVLSLKGGVGKTTITATLGSTFGSARG
MAB_3972c|M.abscessus_ATCC_199 TKKTEENDLIAQVNQPLQGCYKIALLSLKGGVGKTTVTATLGATFASIRG
MLBr_00798|M.leprae_Br4923 AKAEQTNELHRRICAPLADVHKVAFVSAKGGVGKTTITVALGNTMARLRG
MUL_3237|M.ulcerans_Agy99 GRPPRAIPDPKPLSS-HGPAKVVAMCNQKGGVGKTTSTINLGAALAEYDR
: . : :*. . ******** : ** ::.
Mb0543|M.bovis_AF2122/97 DRVVAVDANPDRGTLSQKVP-LETPATVRHLLRDADGIERYSDVRGYTSK
Rv0530|M.tuberculosis_H37Rv DRVVAVDANPDRGTLSQKVP-LETPATVRHLLRDADGIERYSDVRGYTSK
MMAR_0876|M.marinum_M DRVVAVDANPDRGTLSQKVP-LETPATVRHLIRDAEGIERYSDVRGYTSQ
MAV_4615|M.avium_104 DRVVAVDANPDRGTLSQKIP-LETAATVRQLLHDAGTIERYSDVRRYTSK
Mvan_0870|M.vanbaalenii_PYR-1 DRVIAVDANPDRGTLSQKVP-LETPATVRHLLRDAEGIAAYSDVRQYTSQ
Mflv_0048|M.gilvum_PYR-GCK DRVIAVDANPDRGTLSQKVP-VETPATVRHLLRDAEGVSAYSDVRRYTSQ
MSMEG_0975|M.smegmatis_MC2_155 DRVIAVDANPDRGTLSQKVP-LETAATVRHLLRDAEGIERYSDVRAYTSQ
TH_0884|M.thermoresistible__bu DRVVAVDANPDRGTLSQKVP-LQTRATVRDLLRDADRIQTYSDMRAYTSQ
MAB_3972c|M.abscessus_ATCC_199 DRVIAVDANPDRGTLSQKVP-LETPATVRHLLRDANSIQRYSDVRSYTSQ
MLBr_00798|M.leprae_Br4923 DRVIAVDVDADLGDLSARFRERGGPQTNIEHFVSARNAKRYADVRVHTVM
MUL_3237|M.ulcerans_Agy99 -RVLLVDMDP-QGALSAGLG--VAHYELEKTIHNVLVEPRVSVDDVLIHT
**: ** :. * ** . . : .. :
Mb0543|M.bovis_AF2122/97 GPSGLEVLASDSDPASSDAFSADDYTR------TLDILERFYGLVLTDCG
Rv0530|M.tuberculosis_H37Rv GPSGLEVLASDSDPASSDAFSADDYTR------TLDILERFYGLVLTDCG
MMAR_0876|M.marinum_M AKNGFEVLASDTDPAASEAFSADDYTR------TLDILERFYGLVLTDCG
MAV_4615|M.avium_104 GPSGLEVLASETDPAVSEAFSADDYVR------ILDILERFYGLVLTDCG
Mvan_0870|M.vanbaalenii_PYR-1 GPSRLEVLASESDPAVSEAFSAADYCR------AVDVLERFYSVVLTDCG
Mflv_0048|M.gilvum_PYR-GCK GPSRLEVLASESDPAVSEAFSATDYVR------ALNILERFYSMVLTDCG
MSMEG_0975|M.smegmatis_MC2_155 GPSRLEVLASESDPAVSEAFSSEDYLR------TLEVLERFYSLVLTDCG
TH_0884|M.thermoresistible__bu SPSRLEVLASESDPAVSEAFSSDDYTR------TLRVLERFYSLVLTDCG
MAB_3972c|M.abscessus_ATCC_199 GPSRLEVLASESDPAVSEAFSAEDYSR------TLEILEKFYSVVLTDCG
MLBr_00798|M.leprae_Br4923 NNDRLEMLGAQNDPRSTYRLGPDDYET------AMKILENHCNVILLDCG
MUL_3237|M.ulcerans_Agy99 RVKNLDLVPSNIDLSAAEIQLVNEVGREQTLGRALYPVLDRYDYVLVDCQ
. :::: :: * : : : : . :* **
Mb0543|M.bovis_AF2122/97 TGLLHSAMSAVLPRSDVLVVVSSGSIDGARSAAATLDWLQAHGHDDQVRN
Rv0530|M.tuberculosis_H37Rv TGLLHSAMSAVLPRSDVLVVVSSGSIDGARSAAATLDWLQAHGHDDQVRN
MMAR_0876|M.marinum_M TGLLHSAMSAILAKSDILIVVSSGSIDGARSASATLDWLEAHGYEELVRN
MAV_4615|M.avium_104 PGLLHSVMKSVLDKADALVVVSSASIDGARSASATLDWLDAHGHEDLVRN
Mvan_0870|M.vanbaalenii_PYR-1 TGMLHSAMSGVIDKADVLVVISSGSVDGARSASATLDWLDAHGHGDMVGN
Mflv_0048|M.gilvum_PYR-GCK TGMLHSAMSAVIDKADVLVVISSGSADGARSASATLDWLDAHGHDDMVRN
MSMEG_0975|M.smegmatis_MC2_155 TGLIHSAMSAVLSKADVLIVVSSGSVDGARSAAATLDWLDAHGHQDLVRN
TH_0884|M.thermoresistible__bu TGLIHSAMSAVLANADVLIVISSGSVDGARSAAATLDWLDAHGHQDMVRN
MAB_3972c|M.abscessus_ATCC_199 TGLMHSAMSSVLEQADTLVVISSGSVDGARSASATLDWLDAHGHRELVRR
MLBr_00798|M.leprae_Br4923 TPVNGPLFNKIVSTVTGLVVVASDDVRGVEGALATLDWLDAHGYARLLQH
MUL_3237|M.ulcerans_Agy99 PSLGLLTVNG-LACADGVVIPTECEYFSLRGLALLTDTVEKVRDRLNPRL
. : .. : ::: :. . . .. * ::
Mb0543|M.bovis_AF2122/97 SIAVVNAVRPRA--------GKVDVGKVVEHFSRRCRAVRVVPFDPHLEE
Rv0530|M.tuberculosis_H37Rv SIAVVNAVRPRA--------GKVDVGKVVEHFSRRCRAVRVVPFDPHLEE
MMAR_0876|M.marinum_M SIAVINAVRPRS--------SKVDMQKVIDHFARRCRAVRLVPFDPHLEE
MAV_4615|M.avium_104 SIAVINGVRPRP--------GKVDMNKVIDHFSRRCRAVQLVPFDPHLEE
Mvan_0870|M.vanbaalenii_PYR-1 SIAVINGVRRGARKGAGKGTAKVDLNKVVDHFARRCRAVCQVPFDPHLEE
Mflv_0048|M.gilvum_PYR-GCK SIAVINAVRPRGRRGTSRGTAKVDLDKIVDHFTRRCRAVCQVPFDAHLEE
MSMEG_0975|M.smegmatis_MC2_155 SVAVLNAVRPRA--------GKVDLSKVVDHFSRRCRAVRLVPFDPHLEE
TH_0884|M.thermoresistible__bu SIAVINAVRPRS--------GKVDLQKVVDHFSRRCRAVKMVPFDPHLEE
MAB_3972c|M.abscessus_ATCC_199 SIAVINAVRPRS--------GKVDMQKVVDHFSRRCRAVRLVPFDPHLEE
MLBr_00798|M.leprae_Br4923 TVVVLNAIQKTK--------PFVDFGIVENQFRKRVPDFYPMPYDPHLAT
MUL_3237|M.ulcerans_Agy99 DITGILLTRYDP-----------RTVNSREVMARVVERFGDLVFDTVITR
:. : : : : : . : :*. :
Mb0543|M.bovis_AF2122/97 GAEIALDRLRRETREALTELAAVVAAGFP-------------GDPRRCKP
Rv0530|M.tuberculosis_H37Rv GAEIALDRLRRETREALTELAAVVAAGFP-------------GDPRRCKP
MMAR_0876|M.marinum_M GAEIVLDRMKRETREALAELAAVVAEGFG-------------PDQRRSSP
MAV_4615|M.avium_104 GAEIDLDRLRRGTREALTELAAIVADGFP-------------G-AQRNPG
Mvan_0870|M.vanbaalenii_PYR-1 GAEISLDRLRADTRESLLELAAAVADGFP-------------GAR-RTDP
Mflv_0048|M.gilvum_PYR-GCK GAEISLDRLRPQTRESLLELAAAVADGFP-------------GARNRSDP
MSMEG_0975|M.smegmatis_MC2_155 GAEISLDRLKPKTREALLELAAVVAGDFGSDRTGALGGADSAGSRRIADP
TH_0884|M.thermoresistible__bu GAEIDLDQLKPQTRQALLELAAVVADGFP-------------GAQQLQSP
MAB_3972c|M.abscessus_ATCC_199 GAEISLDRLRPQTRRALIELAAVVAQGFT-------------GTQRQQQT
MLBr_00798|M.leprae_Br4923 GLAVEYTSLKRKTRKALKELVGGVAQYYP-----------VIRAQQRDED
MUL_3237|M.ulcerans_Agy99 TVRFPETSVAGEPITTWAPKSAGALAYRA--------------LAREFID
. : . : . .
Mb0543|M.bovis_AF2122/97 SFT--------------
Rv0530|M.tuberculosis_H37Rv SFT--------------
MMAR_0876|M.marinum_M SFG--------------
MAV_4615|M.avium_104 VLG--------------
Mvan_0870|M.vanbaalenii_PYR-1 VG---------------
Mflv_0048|M.gilvum_PYR-GCK AD---------------
MSMEG_0975|M.smegmatis_MC2_155 RV---------------
TH_0884|M.thermoresistible__bu PF---------------
MAB_3972c|M.abscessus_ATCC_199 FQG--------------
MLBr_00798|M.leprae_Br4923 GLGTWIETIRQVEMTSR
MUL_3237|M.ulcerans_Agy99 RFGV-------------