For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VATNGGGAEPTARHQLMTRAAGVGLLMRSTINLGVSLIALADPLSTALPAGRWLLAVLAVWSLYRLATRS HATALLAVDYGLVLAVCLAVPLLVPVTDFHTFNTAPQAIAGTAVISFSVAVRVAACVPMAVGIAAAYAAG AASVIGWENVLSVMALYYFAVQCGTASIIRFMLLRVAGAIDQARRERAEAEVAERVTDAVRDYEREQLAL LHDTAASTLMMVGQGVSLPPERMAAQARRDLHLLRAGSWEAPPPRVELVAALRDCTARLSTPVEFDGRER LWVAGATANPVIAAAREAMNNVDRHARAELLRITVTETCVRLADDGIGFDLGHPRTGHGVDDSIIGRMAR AGGDAHITSSPGAGTVVEMSWAPSAPSAITPPAVDPDRLIDRTRTRYGLALTAYSLVNLAITVPTSDAVL GILATLTSLAAIPGIMWQRWLFAWPAMLAALIVAIGQPAMLPQDHLLGYPHWAQGAIGWCVLPLMLALPT RTGAALVMAYWACNSLVLVIRDPSPLILVNIGYGTASILGIQLFALVFNGLMRDAATAVESENAARQRLL TRDRVSQALRAEYQRRYATIVAGVVPLLEALTRGEKIDTAMQLRARAECRRLRALFDQASTFDHPLMQRI RPLIDAAEGRRVDVAIDVSGTLPDLDDEQISALATPVTEVLNRASTSARIVVTGADGQVEISVVIDIADT ADTPPADVDGAEVIVSGRELWCLIVSGVQKG
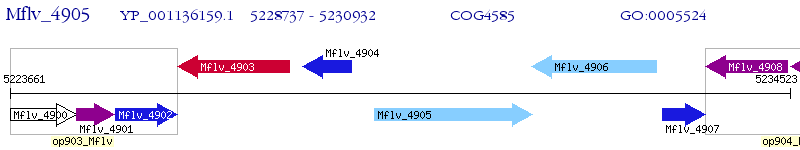
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4905 | - | - | 100% (731) | Signal transduction histidine kinase-like protein |
| M. gilvum PYR-GCK | Mflv_3580 | - | 3e-11 | 27.68% (383) | putative signal transduction histidine kinase |
| M. gilvum PYR-GCK | Mflv_5015 | - | 4e-08 | 30.30% (231) | GAF sensor signal transduction histidine kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3156c | devS | 9e-07 | 25.48% (365) | two component sensor histidine kinase DEVS |
| M. tuberculosis H37Rv | Rv3132c | devS | 9e-07 | 25.48% (365) | two component sensor histidine kinase DEVS |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4520c | - | 5e-12 | 28.21% (390) | putative two-component system sensor kinase |
| M. marinum M | MMAR_3958 | - | 4e-08 | 27.59% (395) | two-component sensor kinase |
| M. avium 104 | MAV_4112 | - | 4e-09 | 30.71% (241) | hypothetical protein MAV_4112 |
| M. smegmatis MC2 155 | MSMEG_3239 | - | 3e-13 | 29.43% (367) | two-component system sensor kinase |
| M. thermoresistible (build 8) | TH_1288 | - | 2e-08 | 26.33% (376) | two-component system sensor kinase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2835 | - | 2e-11 | 28.72% (390) | putative signal transduction histidine kinase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3239|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_2835|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_1288|M.thermoresistible__bu --------------------------------------------------
MMAR_3958|M.marinum_M --------------------------------------------------
MAV_4112|M.avium_104 --------------------------------------------------
MAB_4520c|M.abscessus_ATCC_199 --------------------------------------------------
Mb3156c|M.bovis_AF2122/97 MTTGGLVDENDGAAMRPLRHTLSQLRLHELLVEVQDRVEQIVEGRDRLDG
Rv3132c|M.tuberculosis_H37Rv MTTGGLVDENDGAAMRPLRHTLSQLRLHELLVEVQDRVEQIVEGRDRLDG
Mflv_4905|M.gilvum_PYR-GCK -------MATNGGGAEPTARHQLMTRAAGVGLLMRSTINLGVSLIALADP
MSMEG_3239|M.smegmatis_MC2_155 ---------------------MQREPDPVTPLWRAAQGFR----------
Mvan_2835|M.vanbaalenii_PYR-1 -----------------------MTPDPATPLWRAAQVFR----------
TH_1288|M.thermoresistible__bu --------------------MAAERVDPATPLWRAAQVFR----------
MMAR_3958|M.marinum_M --------------------MATRGPDPTAPLWRAAQLFR----------
MAV_4112|M.avium_104 ----------------MLTTMANHGPDPTTPLWRAAQVFR----------
MAB_4520c|M.abscessus_ATCC_199 -----------MGSKNGTVPRWMSGSDPTASLWRAAQVFR----------
Mb3156c|M.bovis_AF2122/97 LVEAMLVVTAGLDLEATLRAIVHSATSLVDARYGAMEVHDRQHRVLHFVY
Rv3132c|M.tuberculosis_H37Rv LVEAMLVVTAGLDLEATLRAIVHSATSLVDARYGAMEVHDRQHRVLHFVY
Mflv_4905|M.gilvum_PYR-GCK LSTALPAGRWLLAVLAVWSLYRLATRSHATALLAVDYGLVLAVCLAVPLL
. . . .
MSMEG_3239|M.smegmatis_MC2_155 -----------------------LLSCLYALG-FHIAITDDLR-----RP
Mvan_2835|M.vanbaalenii_PYR-1 -----------------------LLSCIYAFG-FQLAVNPDLE-----HP
TH_1288|M.thermoresistible__bu -----------------------LLSCLYAVG-FQIAVNGDLR-----HP
MMAR_3958|M.marinum_M -----------------------LLSCGYALG-FHIAINANLQ-----RP
MAV_4112|M.avium_104 -----------------------LLSCLYATG-FQIAINPDLL-----RP
MAB_4520c|M.abscessus_ATCC_199 -----------------------LASFGYALLASQLASYTNYE-----RQ
Mb3156c|M.bovis_AF2122/97 EGIDEETVRRIGHLPKGLGVIGLLIEDPKPLRLDDVSAHPASIGFPPYHP
Rv3132c|M.tuberculosis_H37Rv EGIDEETVRRIGHLPKGLGVIGLLIEDPKPLRLDDVSAHPASIGFPPYHP
Mflv_4905|M.gilvum_PYR-GCK VPVTDFHTFNTAPQAIAGTAVISFSVAVRVAACVPMAVGIAAAYAAGAAS
: ::
MSMEG_3239|M.smegmatis_MC2_155 VLGWVLFAGLIVW-----------------------------------SA
Mvan_2835|M.vanbaalenii_PYR-1 VGGWVLFAVLIGW-----------------------------------SA
TH_1288|M.thermoresistible__bu AAGWLMLAVLLAW-----------------------------------SA
MMAR_3958|M.marinum_M LLGWALFAVLIGW-----------------------------------SA
MAV_4112|M.avium_104 VLGWVLFAGLIGW-----------------------------------SA
MAB_4520c|M.abscessus_ATCC_199 GLSWALYAVMAMW-----------------------------------TG
Mb3156c|M.bovis_AF2122/97 PMRTFLGVPVRVR-----------------------------------DE
Rv3132c|M.tuberculosis_H37Rv PMRTFLGVPVRVR-----------------------------------DE
Mflv_4905|M.gilvum_PYR-GCK VIGWENVLSVMALYYFAVQCGTASIIRFMLLRVAGAIDQARRERAEAEVA
:
MSMEG_3239|M.smegmatis_MC2_155 ACATVYLTGSRRRGTWVIAELVVVVALMLSTELVASEQWIADNQS-----
Mvan_2835|M.vanbaalenii_PYR-1 ACATAYLQGFGRRPAWVLAEIVVVVALMLSTEFVASDQWALDNQS-----
TH_1288|M.thermoresistible__bu ACAVGYLNGFARRPGWVIAEIVVVIALMLATGLIADAAWVADNQS-----
MMAR_3958|M.marinum_M ACAVAYLRGFGRRPAWVITEIAVVVVLMLSTKMVASAQWALDNQT-----
MAV_4112|M.avium_104 ACALAYLRGFGRKPAWVLAELVVVVLLMWSNHLVASPHWAADNQT-----
MAB_4520c|M.abscessus_ATCC_199 FAAVALSRNWFRRWQLVLCDQAIVIALMLAGRLVVDVQWYSGHQT-----
Mb3156c|M.bovis_AF2122/97 SFGTLYLTDKTNGQPFSDDDEVLVQALAAAAGIAVANARLYQQAK-----
Rv3132c|M.tuberculosis_H37Rv SFGTLYLTDKTNGQPFSDDDEVLVQALAAAAGIAVANARLYQQAK-----
Mflv_4905|M.gilvum_PYR-GCK ERVTDAVRDYEREQLALLHDTAASTLMMVGQGVSLPPERMAAQARRDLHL
. . : . : . . :
MSMEG_3239|M.smegmatis_MC2_155 ---------------------------------WPTTLWATNATISVALH
Mvan_2835|M.vanbaalenii_PYR-1 ---------------------------------WPTTLWATNATISAAIL
TH_1288|M.thermoresistible__bu ---------------------------------LPTTLWATNATISAAIQ
MMAR_3958|M.marinum_M ---------------------------------WPTTLWASNATISAALQ
MAV_4112|M.avium_104 ---------------------------------WPTTLWASNPTISAAIQ
MAB_4520c|M.abscessus_ATCC_199 ---------------------------------LPTTLWVANAVVSMGLL
Mb3156c|M.bovis_AF2122/97 ---------------------------------ARQSWIEATRDIATELL
Rv3132c|M.tuberculosis_H37Rv ---------------------------------ARQSWIEATRDIATELL
Mflv_4905|M.gilvum_PYR-GCK LRAGSWEAPPPRVELVAALRDCTARLSTPVEFDGRERLWVAGATANPVIA
: :
MSMEG_3239|M.smegmatis_MC2_155 FG--------------------------------------PIGGMSAGLA
Mvan_2835|M.vanbaalenii_PYR-1 SG--------------------------------------PVSGMLAGLV
TH_1288|M.thermoresistible__bu FG--------------------------------------PVRGMLTGVA
MMAR_3958|M.marinum_M FG--------------------------------------PAGGMLTGIA
MAV_4112|M.avium_104 FG--------------------------------------PVGGMLTGLA
MAB_4520c|M.abscessus_ATCC_199 RG--------------------------------------QLAGVISGFV
Mb3156c|M.bovis_AF2122/97 SGTEPATVFRLVAAEALKLTAADAALVAVPVD-----EDMPAADVGELLV
Rv3132c|M.tuberculosis_H37Rv SGTEPATVFRLVAAEALKLTAADAALVAVPVD-----EDMPAADVGELLV
Mflv_4905|M.gilvum_PYR-GCK AAREAMNNVDRHARAELLRITVTETCVRLADDGIGFDLGHPRTGHGVDDS
. .
MSMEG_3239|M.smegmatis_MC2_155 VMATVALLKGHVSVNLGRNATIVIELAVG---------------------
Mvan_2835|M.vanbaalenii_PYR-1 VVAASAALKGYVSIDLGRNATVVIELAVG---------------------
TH_1288|M.thermoresistible__bu VMLAAAALKGHISINLGRNATIVIELAVG---------------------
MMAR_3958|M.marinum_M VTAANEAVKGYVNVNLGRSATIVIELAVG---------------------
MAV_4112|M.avium_104 VMTANFAVKNYFALNLGHNATVIIELAIG---------------------
MAB_4520c|M.abscessus_ATCC_199 LAAVSIAARGLPVSALWLDATMPALVVVG---------------------
Mb3156c|M.bovis_AF2122/97 IETVGSAVASTVGRTIPVAGAVLREVFVNGIPRRVDRVDLEGLDELADAG
Rv3132c|M.tuberculosis_H37Rv IETVGSAVASIVGRTIPVAGAVLREVFVNGIPRRVDRVDLEGLDELADAG
Mflv_4905|M.gilvum_PYR-GCK IIGRMARAGGDAHITSSPGAGTVVEMSWAPSAPSAITPPAVDPDRLIDRT
: . . :
MSMEG_3239|M.smegmatis_MC2_155 -----------------------------------LAVGMAAQTARRAHA
Mvan_2835|M.vanbaalenii_PYR-1 -----------------------------------LAVGMAAQTARRAHA
TH_1288|M.thermoresistible__bu -----------------------------------LAVGMAAQTARRAHA
MMAR_3958|M.marinum_M -----------------------------------LAVGMAARTARRAHA
MAV_4112|M.avium_104 -----------------------------------MAIGMAAQTARRAHD
MAB_4520c|M.abscessus_ATCC_199 -----------------------------------LAIGAASTTAKRTFH
Mb3156c|M.bovis_AF2122/97 PALLLPLRARGTVAGVVVVLSQGGPGAFTDEQLEMMAAFADQAALAWQLA
Rv3132c|M.tuberculosis_H37Rv PALLLPLRARGTVAGVVVVLSQGGPGAFTDEQLEMMAAFADQAALAWQLA
Mflv_4905|M.gilvum_PYR-GCK RTRYGLALTAYSLVNLAITVPTSDAVLGILATLTSLAAIPGIMWQRWLFA
:*
MSMEG_3239|M.smegmatis_MC2_155 ELERAVRLSAALEERERLSR-------------------------RVHDG
Mvan_2835|M.vanbaalenii_PYR-1 ELERAARLSASLEERERLSR-------------------------QVHDG
TH_1288|M.thermoresistible__bu ELRRAARLAAVTEERERLSR-------------------------QVHDG
MMAR_3958|M.marinum_M DLQRAAQLSAALQERERLSR-------------------------QVHDG
MAV_4112|M.avium_104 ELQRAARLTAAAQERDRLAR-------------------------QVHDG
MAB_4520c|M.abscessus_ATCC_199 ELRRAERAAAAAIERERLSR-------------------------QVHDG
Mb3156c|M.bovis_AF2122/97 TSQRRMRELDVLTDRDRIAR-------------------------DLHDH
Rv3132c|M.tuberculosis_H37Rv TSQRRMRELDVLTDRDRIAR-------------------------DLHDH
Mflv_4905|M.gilvum_PYR-GCK WPAMLAALIVAIGQPAMLPQDHLLGYPHWAQGAIGWCVLPLMLALPTRTG
: :.: :
MSMEG_3239|M.smegmatis_MC2_155 AIQVLALV---------ARRGREIGG-------ETAKLAELAGEQERALR
Mvan_2835|M.vanbaalenii_PYR-1 AIQVLALV---------ARRGREIGG-------DTAQLAELAGEQERALR
TH_1288|M.thermoresistible__bu AIQVLALV---------ARRGREIGG-------EVTELAELAGEQERALR
MMAR_3958|M.marinum_M VIQVLALV---------SRRGHEIGG-------PTSELAELASEQERALR
MAV_4112|M.avium_104 AIQVLALV---------AKKGHEIGG-------ATTELADLASEQERALR
MAB_4520c|M.abscessus_ATCC_199 VLQVLSYV---------KRRGTEIGG-------PTAELAAIAGEQEIALR
Mb3156c|M.bovis_AF2122/97 VIQRLFAIGLALQGAVPHERNPEVQQ-------RLSDVVDDLQDVIQEIR
Rv3132c|M.tuberculosis_H37Rv VIQRLFAIGLALQGAVPHERNPEVQQ-------RLSDVVDDLQDVIQEIR
Mflv_4905|M.gilvum_PYR-GCK AALVMAYWACNSLVLVIRDPSPLILVNIGYGTASILGIQLFALVFNGLMR
. : . : : :*
MSMEG_3239|M.smegmatis_MC2_155 RLVSAADTDTM-AGPLTDVGALLRTRASD---RVSVSVPAEPVLLDHPV-
Mvan_2835|M.vanbaalenii_PYR-1 RLVSANDAEPR-KGAMSDIGAMLRRRATD---RVSISVPADPVLLDATV-
TH_1288|M.thermoresistible__bu RLVSATESAPGGADHTVDVATLLRRRACD---RVTMSLPGTPVPMDADV-
MMAR_3958|M.marinum_M RWIISTDTDRDGNTRSVDLCALLRRRASD---RVSMSLPGTPVTLTREV-
MAV_4112|M.avium_104 RWLACTDIDRDADGDTVDLRTLLRRRESD---RVSISLPGTPVRLGRWA-
MAB_4520c|M.abscessus_ATCC_199 ELISGATQTTLADDELVDLRPLLRTQAST---TVSVSAPGAPVLIGQHM-
Mb3156c|M.bovis_AF2122/97 TTIYDLHGASQGITRLRQRIDAAVAQFADSGLRTSVQFVGPLSVVDSAL-
Rv3132c|M.tuberculosis_H37Rv TTIYDLHGASQGITRLRQRIDAAVAQFADSGLRTSVQFVGPLSVVDSAL-
Mflv_4905|M.gilvum_PYR-GCK DAATAVESENAARQRLLTRDRVSQALRAEYQRRYATIVAGVVPLLEALTR
: . :
MSMEG_3239|M.smegmatis_MC2_155 -----------ARELFAAAENALDNVAAHAGADARAFVLLEDLGEEVTVS
Mvan_2835|M.vanbaalenii_PYR-1 -----------ADELLAAVTNALDNVEAHAGPVARAYVLLEDLGATVTVT
TH_1288|M.thermoresistible__bu -----------AAELDAAVGNALDNVALHAGPDARAFVLVEDLGDTVTVS
MMAR_3958|M.marinum_M -----------ATELDAAVGNALDNVRLHAGPSARAFVLLEDLGDSVTVS
MAV_4112|M.avium_104 -----------ATELDAAVGNALDNVVAHAGPGAHAFVLVEDLGDSVLVS
MAB_4520c|M.abscessus_ATCC_199 -----------ASELAAVVRSALSNVALHAGPQAHAYVLIEDLVDSVVVT
Mb3156c|M.bovis_AF2122/97 -----------ADQAEAVVREAVSNAVRHA--KASTLTVRVKVDDDLCIE
Rv3132c|M.tuberculosis_H37Rv -----------ADQAEAVVREAVSNAVRHA--KASTLTVRVKVDDDLCIE
Mflv_4905|M.gilvum_PYR-GCK GEKIDTAMQLRARAECRRLRALFDQASTFDHPLMQRIRPLIDAAEGRRVD
* ..:. . . :
MSMEG_3239|M.smegmatis_MC2_155 IRDDGVGIPEGRLAEAERQGRMGVAKSIVGRMDWLGGTAVLTTGP-DSGT
Mvan_2835|M.vanbaalenii_PYR-1 VRDDGVGIPEGRLDEAVTEGRVGVSKSIVGRMDWLGGRAELSTGP-GMGT
TH_1288|M.thermoresistible__bu IRDDGAGIAPGRLDEAVREGRMGIAKSIVGRMESLGGEAVLTTS--ERGT
MMAR_3958|M.marinum_M VRDDGVGIAPGRLDEAAGQGHLGISKSIVGRLAALGGTAELSTDQ-GEGT
MAV_4112|M.avium_104 VRDDGVGIAAGRVEEAARQGRLGISQSIVGRLASLGGTAELHSEP-GAGT
MAB_4520c|M.abscessus_ATCC_199 VRDDGSGIADGRLDQAEREGRMGVSRSILGRVADLGGAVLLETGP-GAGT
Mb3156c|M.bovis_AF2122/97 VTDNGRGLP----DEFTGSG----LTNLRQRAEQAGGEFTLASVPGASGT
Rv3132c|M.tuberculosis_H37Rv VTDNGRGLP----DEFTGSG----LTNLRQRAEQAGGEFTLASVPGASGT
Mflv_4905|M.gilvum_PYR-GCK VAIDVSGTLPDLDDEQISALATPVTEVLNRASTSARIVVTGADGQVEISV
: : * : : ..
MSMEG_3239|M.smegmatis_MC2_155 EWELTVPRTRKGQ-------------------------
Mvan_2835|M.vanbaalenii_PYR-1 EWELTVSR------------------------------
TH_1288|M.thermoresistible__bu EWELTVPRKDGRHG------------------------
MMAR_3958|M.marinum_M EWELRVPRREGSS-------------------------
MAV_4112|M.avium_104 EWELCVPRREGRDDG-----------------------
MAB_4520c|M.abscessus_ATCC_199 EWEITVPKSQEVGA------------------------
Mb3156c|M.bovis_AF2122/97 VLRWSAPLSQ----------------------------
Rv3132c|M.tuberculosis_H37Rv VLRWSAPLSQ----------------------------
Mflv_4905|M.gilvum_PYR-GCK VIDIADTADTPPADVDGAEVIVSGRELWCLIVSGVQKG
.