For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTRPLLTADRELALLRELIQAASSGPGVEPLAAASARMITASTDSDVCFVHVLDDTERSLTLAGATPPFD SEVGKIRLPLGQGISGWVASHRQPVVIRRDKESDPRYLPFQSLRGRDFTSMVSVPMETDPGGLVGVLNVH TVAERDFTDRDVELLLVIGRLIAGAMHQARLHRQLVARERAHENFVEQVIEAQELERRRLAGDIHDGISQ RLVTLSYRLDAAAQAAKSPADHAALTEQLDRARELADLTLQEARAAISGLRPPVLDDLGLAGGLASLARS IPQIGIETDLDDTRLADHIELALYRIAQECLQNVVKHARASAARLTFAVDDNTARLEIADDGVGFDTFEH PLGSDEMGGYGLLSMAERAEIVGGRLNIRSRPGSGTTVTATIPLHQGGA
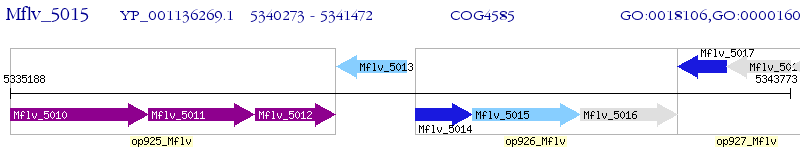
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5015 | - | - | 100% (399) | GAF sensor signal transduction histidine kinase |
| M. gilvum PYR-GCK | Mflv_0729 | - | 2e-22 | 32.46% (228) | integral membrane sensor signal transduction histidine kinase |
| M. gilvum PYR-GCK | Mflv_2281 | - | 2e-20 | 29.07% (258) | integral membrane sensor signal transduction histidine kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2052c | - | 1e-18 | 27.40% (292) | histidine kinase response regulator |
| M. tuberculosis H37Rv | Rv2027c | - | 1e-18 | 27.40% (292) | histidine kinase response regulator |
| M. leprae Br4923 | MLBr_00774 | mtrB | 3e-07 | 22.98% (248) | putative two-component system sensor kinase |
| M. abscessus ATCC 19977 | MAB_4499 | - | 1e-28 | 33.08% (263) | putative two-component system sensor kinase |
| M. marinum M | MMAR_4781 | - | 3e-18 | 32.90% (231) | two-component sensor kinase |
| M. avium 104 | MAV_0874 | - | 6e-19 | 31.90% (232) | hypothetical protein MAV_0874 |
| M. smegmatis MC2 155 | MSMEG_1493 | - | 0.0 | 85.46% (392) | sensor histidine kinase |
| M. thermoresistible (build 8) | TH_4203 | - | 0.0 | 82.84% (402) | PUTATIVE sensor histidine kinase |
| M. ulcerans Agy99 | MUL_0350 | - | 2e-18 | 32.90% (231) | two component sensor kinase |
| M. vanbaalenii PYR-1 | Mvan_1357 | - | 0.0 | 90.95% (398) | GAF sensor signal transduction histidine kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2052c|M.bovis_AF2122/97 -----------------------------------------MTHPDRANV
Rv2027c|M.tuberculosis_H37Rv -----------------------------------------MTHPDRANV
Mflv_5015|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1357|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1493|M.smegmatis_MC2_155 --------------------------------------------------
TH_4203|M.thermoresistible__bu --------------------------------------------------
MMAR_4781|M.marinum_M --------------------------------------------------
MUL_0350|M.ulcerans_Agy99 MMNSRQREADRRQFAPSCRRQDESAGQSLGQRRTERSPNPSCANYGQLVG
MAV_0874|M.avium_104 --------------------------------------------------
MAB_4499|M.abscessus_ATCC_1997 ------------MQQAAQASSPRPILMPDYISGWRTQLFGSTPPSARLGI
MLBr_00774|M.leprae_Br4923 ------MIFSSRRRIRGRWGRSGPMMRGMGALTRVVGVVWRRSLQLRVVA
Mb2052c|M.bovis_AF2122/97 NPGSPPVRETLSQLRLRELLLEVQDRIEQIVEGRDRLDGLIDAILAITSG
Rv2027c|M.tuberculosis_H37Rv NPGSPPLRETLSQLRLRELLLEVQDRIEQIVEGRDRLDGLIDAILAITSG
Mflv_5015|M.gilvum_PYR-GCK -------------------------MTRPLLTADRELALLRELIQAASSG
Mvan_1357|M.vanbaalenii_PYR-1 -------------------------MSRPLLTADRELALLRELIQAASSG
MSMEG_1493|M.smegmatis_MC2_155 ------------MNAAKRAVKPVRDL----VDADRELALLRELIQAASSG
TH_4203|M.thermoresistible__bu -------------VTSGPHPDPVRDLSEQLLTADRELVLLRELIQAASRG
MMAR_4781|M.marinum_M ---------MVTSDAELARVRRTHQLRAYRIGSVLRIGVVLFMVAAMLVG
MUL_0350|M.ulcerans_Agy99 SCRLYGVPVVVTSDAELARVRRTHQLRAYRIGSVLRIGVVLFMVAAMLVG
MAV_0874|M.avium_104 --------MAVAGDAELERVRAVHQMRSYRIGSVLRVGVVGLMVAAMIIG
MAB_4499|M.abscessus_ATCC_1997 VAAVVLLVAESMLTIFLRTIAPTEHLAAVYLIGILVISVIWRLRLALAMS
MLBr_00774|M.leprae_Br4923 LTFGLSLAVILALGFVLTSQLTSRVLDVKVRVAIEQIERARTTVTGIVNG
. : . .
Mb2052c|M.bovis_AF2122/97 LKLDATLRAI-----VHTAAELVDARYGALGVRGYDHRLVEFVYEGIDEE
Rv2027c|M.tuberculosis_H37Rv LKLDATLRAI-----VHTAAELVDARYGALGVRGYDHRLVEFVYEGIDEE
Mflv_5015|M.gilvum_PYR-GCK PGVEPLAAAS-----ARMITASTDSDVCFVHVLDDTERSLTLAGATPPFD
Mvan_1357|M.vanbaalenii_PYR-1 PGVEPLAAAA-----ARMITAATDSDVCFVHVLDDTERSLTLAGATPPFD
MSMEG_1493|M.smegmatis_MC2_155 PGVEPLAAAA-----ARMITAATGTDVCFVHVLDDSDRSLTLAGATPPFD
TH_4203|M.thermoresistible__bu PGVEPLAAAA-----ARMITAATASDVCFVHVLDDTERSLTLAGATPPFD
MMAR_4781|M.marinum_M TPRREWGAQT-----ALICVYAFAALWALALAFRRSRPMVSLDRFANIAR
MUL_0350|M.ulcerans_Agy99 TPRREWGAQT-----ALICVYAFAALWALALAFRRSRPMVSLDRFANIAR
MAV_0874|M.avium_104 TARSEWPQES-----VLILLYGIIALGAVALAFAPFRRWIGIGRLAGVGR
MAB_4499|M.abscessus_ATCC_1997 IASAIVFDCIRNWPKANPLSTESHNLITHLSFLAVTLTASALAALARAGT
MLBr_00774|M.leprae_Br4923 EETRSLDSSLQLARNTLTSKTDPTSGAGLVGAFDAVLIVPGDGPRTATTA
.
Mb2052c|M.bovis_AF2122/97 TRHLIGSLPEGRGVLGALIEEPKPIRLD-DISRHPASVGFP-LHHPP-MR
Rv2027c|M.tuberculosis_H37Rv TRHLIGSLPEGRGVLGALIEEPKPIRLD-DISRHPASVGFP-LHHPP-MR
Mflv_5015|M.gilvum_PYR-GCK SEVGKIRLPLGQGISGWVASHRQPVVIRRDKESDPRYLPFQSLRGRD-FT
Mvan_1357|M.vanbaalenii_PYR-1 SEVGKIRLPLGQGISGWVASNREPVVIRRDKESDPRYLPFRSLRGRD-FT
MSMEG_1493|M.smegmatis_MC2_155 SEVGKIRLPIGQGISGWVADHCEPVVISHDKEADPRYMPFESLRGRD-FT
TH_4203|M.thermoresistible__bu KEVGKIRLPLGQGISGWVASHREPVVITENKEADPRYLPFQSLRGRD-FT
MMAR_4781|M.marinum_M YEPLAFTAVDVLALTGFQLLSTDGIYSLLIMTLLPVLVALDISSRRA-AV
MUL_0350|M.ulcerans_Agy99 YEPLAFTAVDVLALTGFQLLSTDGIYSLLIMTLLPVLVALDISSRRA-AV
MAV_0874|M.avium_104 LEPFAFTVVDILALTIFQLLSTNGIYPLLIMAMLPVLLGLDVSSRRA-AV
MAB_4499|M.abscessus_ATCC_1997 IEAERRRREAAAIAQQQAALRRTATLVASNVSPVELYAKVVEEAARC-FA
MLBr_00774|M.leprae_Br4923 GPVDQVPNSLRGFIKAGQAAYQYATVHTEGFSGPALIIGTPTSSQVTNLE
Mb2052c|M.bovis_AF2122/97 TFLGVPVRIRDEVFGNLYLTEKADGQPFSDDDEVLVQALAAAAGIAV-DN
Rv2027c|M.tuberculosis_H37Rv TFLGVPVRIRDEVFGNLYLTEKADGQPFSDDDEVLVQALAAAAGIAV-DN
Mflv_5015|M.gilvum_PYR-GCK SMVSVPMETDPGGLVGVLNVHTVAERDFTDRDVELLLVIGRLIAGAM-HQ
Mvan_1357|M.vanbaalenii_PYR-1 SMVSVPMETDPGGLVGVLNVHTVAERDFGDRDVGLLLVIGRLIAGAM-HQ
MSMEG_1493|M.smegmatis_MC2_155 SMVSVPMEAEPGGLVGVLNVHTVRRREFTPRDVELLVVIGRLIAGAL-HQ
TH_4203|M.thermoresistible__bu SMVSVPMETDPGGLVGVLNVHTVERREFTRRDVDLLLVIGRLIAGGL-HQ
MMAR_4781|M.marinum_M VLALTLVGFALALAQDPVVIRMNGWPEAIFRFGLYAFLCATALMVVR-SE
MUL_0350|M.ulcerans_Agy99 VLALTLVGFGLALAQDPVVIRMNGWPEAIFRFGLYAFLCATALMVVR-SE
MAV_0874|M.avium_104 VLIFSMLGFAIAVLQDPVMSSSVRLPEAGFRFLLYGFLCCAAFLVVR-IE
MAB_4499|M.abscessus_ATCC_1997 SPTAVLVQLMPGDEGTIVAGHGTEESRTPPAGKRFPLLDSNNVARELRLT
MLBr_00774|M.leprae_Br4923 LYLIFPLKNEQATVTLVRGTMATGGMVLLVLLSGIALLVSRQVVVPVRSA
:
Mb2052c|M.bovis_AF2122/97 ARLFEESRTREAWIEATRDIGTQMLAGADPAMVFRLIAEEALTLMAGAAT
Rv2027c|M.tuberculosis_H37Rv ARLFEESRTREAWIEATRDIGTQMLAGADPAMVFRLIAEEALTLMAGAAT
Mflv_5015|M.gilvum_PYR-GCK ARLHRQLVARERAHENFVEQ------------------------------
Mvan_1357|M.vanbaalenii_PYR-1 ARLHRQLVARERAHEHFVEQ------------------------------
MSMEG_1493|M.smegmatis_MC2_155 ARLHRQLVARERAHENFVEQ------------------------------
TH_4203|M.thermoresistible__bu ARLHRQLVARERAHEIFVEQ------------------------------
MMAR_4781|M.marinum_M ERHARSVATLSVIREELLAQ------------------------------
MUL_0350|M.ulcerans_Agy99 ERHARSVATLSVIREELLAQ------------------------------
MAV_0874|M.avium_104 ERHTRSVAGLSALREELLAQ------------------------------
MAB_4499|M.abscessus_ATCC_1997 PAVCAPIVVDAKHWGTLIVGSHTPAQRDQDIRAGVRDFADLVATAIANAA
MLBr_00774|M.leprae_Br4923 SRIAERFAEGHLSERMPVRG------------------EDDMARLAVSFN
Mb2052c|M.bovis_AF2122/97 LVAVPLDDEAPACEVDDLVIVEVAGEISPAVKQMTVAVSGTSIGGVFHDR
Rv2027c|M.tuberculosis_H37Rv LVAVPLDDEAPACEVDDLVIVEVAGEISPAVKQMTVAVSGTSIGGVFHDR
Mflv_5015|M.gilvum_PYR-GCK -----------VIEAQELERRRLAGDIHDGISQRLVTLS-----------
Mvan_1357|M.vanbaalenii_PYR-1 -----------VIEAQELERRRLAGDIHDGISQRLVTLS-----------
MSMEG_1493|M.smegmatis_MC2_155 -----------VIEAQELERRRLAADIHDGISQRLVTLS-----------
TH_4203|M.thermoresistible__bu -----------VIEAQELERRRLAADIHDGISQRLVTLS-----------
MMAR_4781|M.marinum_M -----------TMTASEVLQRRISESIHDGPLQDVLAAR-----------
MUL_0350|M.ulcerans_Agy99 -----------TMTASEVLQRRISESIHDGPLQDVLAAR-----------
MAV_0874|M.avium_104 -----------TMNASDVSQRRISEFIHDGPLQDILAVR-----------
MAB_4499|M.abscessus_ATCC_1997 TREELAASRARIVSASDTARRRLERDLHDGAQQRLAYLR-----------
MLBr_00774|M.leprae_Br4923 DMAESLSRQITQLEEFGNLQRRFTSDVSHELRTPLTTVR---------MA
.. :
Mb2052c|M.bovis_AF2122/97 TPRRFDRLDLAVDGPVEPGPALVLPLRAADTVAGVLVALRSADEQPFSDK
Rv2027c|M.tuberculosis_H37Rv TPRRFDRLDLAVDGPVEPGPALVLPLRAADTVAGVLVALRSADEQPFSDK
Mflv_5015|M.gilvum_PYR-GCK --YRLDAAAQAAKSPADHAALTEQLDRARELADLTLQEARAAISG-----
Mvan_1357|M.vanbaalenii_PYR-1 --YRLDAAAQAATSPGDPDALHEQLDRARELAELTLQEARAAISG-----
MSMEG_1493|M.smegmatis_MC2_155 --YRLDAAARAAD---DPRAVAEQLGKARELADLTLQEARAAISG-----
TH_4203|M.thermoresistible__bu --YRLDAAIRAVD---DPATVTEQLSKARELADLTLEETRAAISG-----
MMAR_4781|M.marinum_M --QELIELRAAAPHDQRVSTALAGLQSASDLLRQATFEL-----------
MUL_0350|M.ulcerans_Agy99 --QELIELRAAAPHDQRVSTALAGLQSASDLLRQATFEL-----------
MAV_0874|M.avium_104 --QELVELDAAVPDDERIGRALAGLQLASERLRQATFEL-----------
MAB_4499|M.abscessus_ATCC_1997 --LKLGMVRAAVP--ATDAELTQELNNLDTDLTAVTRELQEFSRG-----
MLBr_00774|M.leprae_Br4923 ADLIYDHSSDLDPTLRRSTELMVSELDRFETLLNDLLEISRHDAG-----
Mb2052c|M.bovis_AF2122/97 QLDMMAAFADQAALAWRLATAQRQMREVEILT--DRDRIARDLHDHVIQR
Rv2027c|M.tuberculosis_H37Rv QLDMMAAFADQAALAWRLATAQRQMREVEILT--DRDRIARDLHDHVIQR
Mflv_5015|M.gilvum_PYR-GCK ---LRPPVLDDLGLAGGLASLARSIPQIGI----ETDLDDTRLADHIELA
Mvan_1357|M.vanbaalenii_PYR-1 ---LRPPVLDDLGLSGGLASLARTIPQVTI----DVDLVDTRLPDHIELA
MSMEG_1493|M.smegmatis_MC2_155 ---LRPPVLDDLGLSGGLASLARSIPQVDL----DVDLADTRVPDHIELA
TH_4203|M.thermoresistible__bu ---LRPPVLDDLGLSGGLASLAQSVGQLDIGVDIEVDLAETRLPDHIELA
MMAR_4781|M.marinum_M ----HPAVLEQVGLGAAVEQLASFHAQRSGIR--ITDDVDYPIRDEIDPI
MUL_0350|M.ulcerans_Agy99 ----HPAVLEQVGLGAAVEQLASFHAQRSGIR--ITDDVDYPIRDEIDPI
MAV_0874|M.avium_104 ----HPAVLEQVGLGAAVQQLAEFTAQRSGIE--ISTDIDYSGRSAIDAV
MAB_4499|M.abscessus_ATCC_1997 --------LHPAILSKGLAPALRTLARRSLIPVRTELELDQKYGESIEIA
MLBr_00774|M.leprae_Br4923 ---VAELSVEAVDLRVMVNNALGNVGHLAEEAG-IELLVDMPVDEVIAEV
. * : . . :
Mb2052c|M.bovis_AF2122/97 LFAVGLTLQGAAPRARVPAVRESIYSSIDDLQEIIQEIRSAIFDLHAGPS
Rv2027c|M.tuberculosis_H37Rv LFAVGLTLQGAAPRARVPAVRESIYSSIDDLQEIIQEIRSAIFDLHAGPS
Mflv_5015|M.gilvum_PYR-GCK LYRI----------------------AQECLQNVVKHAR-----------
Mvan_1357|M.vanbaalenii_PYR-1 LYRI----------------------AQECLQNVVKHAR-----------
MSMEG_1493|M.smegmatis_MC2_155 LYRI----------------------AQECLQNVVKHAR-----------
TH_4203|M.thermoresistible__bu LYRI----------------------AQECLQNVVKHAR-----------
MMAR_4781|M.marinum_M VFGV----------------------ARELLSNVVQHSR-----------
MUL_0350|M.ulcerans_Agy99 VFGV----------------------ARELLSNVVQHSR-----------
MAV_0874|M.avium_104 VFGV----------------------VRELLSNVVQHSR-----------
MAB_4499|M.abscessus_ATCC_1997 AYYV----------------------VAEALANTAKHSQ-----------
MLBr_00774|M.leprae_Br4923 DARR----------------------VERILRNLIANAID----------
* : .
Mb2052c|M.bovis_AF2122/97 RATGLRHRLDKVIDQLAIPALHTTVQYTGPLSVVDTVLANHAEAVLREAV
Rv2027c|M.tuberculosis_H37Rv RATGLRHRLDKVIDQLAIPALHTTVQYTGPLSVVDTVLANHAEAVLREAV
Mflv_5015|M.gilvum_PYR-GCK --------------------------------------------ASAARL
Mvan_1357|M.vanbaalenii_PYR-1 --------------------------------------------ASRARL
MSMEG_1493|M.smegmatis_MC2_155 --------------------------------------------ATTARL
TH_4203|M.thermoresistible__bu --------------------------------------------ARTARL
MMAR_4781|M.marinum_M --------------------------------------------ARNASV
MUL_0350|M.ulcerans_Agy99 --------------------------------------------ARNASV
MAV_0874|M.avium_104 --------------------------------------------ARTAAI
MAB_4499|M.abscessus_ATCC_1997 -----------------------------------------------AS-
MLBr_00774|M.leprae_Br4923 ---------------------------------------------HSEHK
Mb2052c|M.bovis_AF2122/97 SNAVRHANATSLAINVSVEDDVRVEVVDD------------GVGISGDIT
Rv2027c|M.tuberculosis_H37Rv SNAVRHANATSLAINVSVEDDVRVEVVDD------------GVGISGDIT
Mflv_5015|M.gilvum_PYR-GCK TFAVD----DNTARLEIADDGVGFDTFEH------------PLGSD-EMG
Mvan_1357|M.vanbaalenii_PYR-1 TFALDRGDHGDLARLEIADDGIGFDTLEH------------PLGGD-EMG
MSMEG_1493|M.smegmatis_MC2_155 TFAVEDG----IARLEIVDDGIGFDTFEH------------PLGSD-EMS
TH_4203|M.thermoresistible__bu TYAVDRGEHGEVARLEIVDDGVGFDTFEH------------PLGGD-DMS
MMAR_4781|M.marinum_M ALGISDG----VCVLDVADDGVGVSGETM------------ARRLG--QG
MUL_0350|M.ulcerans_Agy99 ALGISDG----VCVLDVADDGVGVSGETM------------ARRLG--QG
MAV_0874|M.avium_104 MLGRTDG----TCVLHVVDDGVGFNQETV------------ARRLG--EG
MAB_4499|M.abscessus_ATCC_1997 ---------EVVIRAHEADDQIHITISDD------------GIGGAKPRK
MLBr_00774|M.leprae_Br4923 PVRIRMAADEDTVAVTVRDYGIGLRPGEEKLVFSRFWRSDPSRVRRSGGT
: : .
Mb2052c|M.bovis_AF2122/97 ESGLRNLRQRADDAGGEFTVENMPTGGTLLRWSAPLR-------------
Rv2027c|M.tuberculosis_H37Rv ESGLRNLRQRADDAGGEFTVENMPTGGTLLRWSAPLR-------------
Mflv_5015|M.gilvum_PYR-GCK GYGLLSMAERAEIVGGRLNIRSRPGSGTTVTATIPLHQGGA---------
Mvan_1357|M.vanbaalenii_PYR-1 GYGLLSMAERAEIVGGRLNIRSRPGAGTTVTATIPLPANRE---------
MSMEG_1493|M.smegmatis_MC2_155 GYGLLSMAERAEIVGGRLNIRSRPGAGTAVTATIPLPSHEPAPNLTKLP-
TH_4203|M.thermoresistible__bu GYGLLSMAERAEIVGGRLNIRSRPGAGTAVIATIPLP---PATR------
MMAR_4781|M.marinum_M HIGLASHRARVDAAGGSLVFLNTP-TGTHVCVRIPLKG------------
MUL_0350|M.ulcerans_Agy99 HIGLASHRARVDAAGGSLVFLNTP-TGTHVCVRIPLKG------------
MAV_0874|M.avium_104 HIGLASHRARVEAAGGELVFLDVP-VGTHVCVEVPLRN------------
MAB_4499|M.abscessus_ATCC_1997 GTGLIGLTDRVTALGGRLDVHSASGRGTTMTAIIPLHQSD----------
MLBr_00774|M.leprae_Br4923 GLGLAISIEDARLHQGRLEAWGEPGQGACFRLTLPLVRGHKVTTSPLPMK
** . * : . . *: . **
Mb2052c|M.bovis_AF2122/97 -------------------------------
Rv2027c|M.tuberculosis_H37Rv -------------------------------
Mflv_5015|M.gilvum_PYR-GCK -------------------------------
Mvan_1357|M.vanbaalenii_PYR-1 -------------------------------
MSMEG_1493|M.smegmatis_MC2_155 -------------------------------
TH_4203|M.thermoresistible__bu -------------------------------
MMAR_4781|M.marinum_M -------------------------------
MUL_0350|M.ulcerans_Agy99 -------------------------------
MAV_0874|M.avium_104 -------------------------------
MAB_4499|M.abscessus_ATCC_1997 -------------------------------
MLBr_00774|M.leprae_Br4923 PILQPSPQASTAGQQHGTQRQRLREHAERSR