For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STRWQQSDRPRGEDYDARWKRLAASGASVHGEADLIEALLREGGGTRVLDAGCGTGRVAIELAARGFDVV GLDADPTMLETARAKAPRLRWIEADLVDTDDHLDETFDVVALPGNVMIFLDRGTEAAVVAALARRLAPGG LLVAGFQLGTGRLTLDRYDEITATAALELVDRWATWDRQPYDGGDYAVSLHRLT*
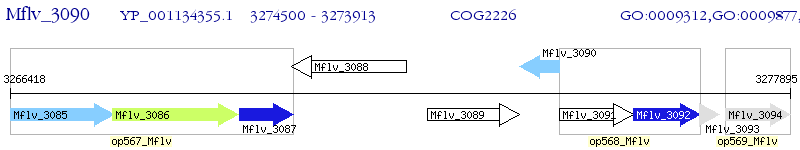
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3090 | - | - | 100% (195) | methyltransferase type 11 |
| M. gilvum PYR-GCK | Mflv_4890 | - | 9e-22 | 35.18% (199) | methyltransferase type 12 |
| M. gilvum PYR-GCK | Mflv_4471 | - | 1e-09 | 38.21% (123) | methyltransferase type 11 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3374 | - | 3e-07 | 36.36% (99) | methyltransferase (methylase) |
| M. tuberculosis H37Rv | Rv3342 | - | 3e-07 | 36.36% (99) | methyltransferase (methylase) |
| M. leprae Br4923 | MLBr_01713 | - | 5e-07 | 31.31% (99) | hypothetical protein MLBr_01713 |
| M. abscessus ATCC 19977 | MAB_2548c | - | 2e-25 | 38.58% (197) | hypothetical protein MAB_2548c |
| M. marinum M | MMAR_1164 | - | 7e-24 | 36.22% (196) | methyltransferase |
| M. avium 104 | MAV_4332 | - | 1e-21 | 34.67% (199) | SAM-dependent methyltransferases |
| M. smegmatis MC2 155 | MSMEG_3430 | - | 2e-21 | 33.50% (197) | SAM-dependent methyltransferase |
| M. thermoresistible (build 8) | TH_1247 | - | 2e-22 | 33.85% (192) | SAM-dependent methyltransferase |
| M. ulcerans Agy99 | MUL_2851 | - | 5e-24 | 36.22% (196) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1534 | - | 3e-24 | 36.55% (197) | methyltransferase type 12 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3430|M.smegmatis_MC2_155 MVEQSLWMQKVAADPGHSQWYIERFRSLARAGED-----------LFGEA
TH_1247|M.thermoresistible__bu MVEKSLWMRKVEADPGHSQWYIERFRAMARAGED-----------LVGEA
MMAR_1164|M.marinum_M MVEQSIWMQKVAADPGHSRWYIERFRAMARAGED-----------LVGEA
MUL_2851|M.ulcerans_Agy99 MVEQSIWMQKVAADPGHSRWYIERFRAMARAGED-----------LVGEA
MAV_4332|M.avium_104 MVEQSIWMQKVAADPGHSHWYIERFRAMARAGDD-----------LAGEA
Mvan_1534|M.vanbaalenii_PYR-1 MVETSIWMQKVAADPGHSRWYIERFRAMARAGDD-----------LAGEA
MAB_2548c|M.abscessus_ATCC_199 MVEQSLWMQKVAADPNHSQWYIDRFRSMARDGAD-----------LAGEA
Mflv_3090|M.gilvum_PYR-GCK --MSTRWQQ---SDRPRGEDYDARWKRLAASGAS-----------VHGEA
Mb3374|M.bovis_AF2122/97 ---------MTCSRRDMSLSFGSAVGAYERGRPS-----------YPPEA
Rv3342|M.tuberculosis_H37Rv ---------MTCSRRDMSLSFGSAVGAYERGRPS-----------YPPEA
MLBr_01713|M.leprae_Br4923 MSAFVPDVPNRIPDDPSSIDSTAFHGMLTLTGERTIPDLDIENYWFRRHQ
. . .
MSMEG_3430|M.smegmatis_MC2_155 RLVDAMAPRN--AHILDAGCGPGRLGGYLAGVGHRVVGVDVDPALIAAAE
TH_1247|M.thermoresistible__bu RLVDAMAPRN--AHILDAGCGPGRLGGYLSAAGHRVVGVDVDPALIEAAE
MMAR_1164|M.marinum_M RLVDSIAPRG--AHILDAGCGPGRLGRHLAAAGHRVVGVDVDPALIEAAE
MUL_2851|M.ulcerans_Agy99 RLVDSIAPRG--AHILDAGCGPGRLGRHLAAAGHRVVGVDVDPALIEAAE
MAV_4332|M.avium_104 RFVDAMAPRG--ARILDAGCGPGRLGSYLAAAGHQVVGVDVDPALIEAAE
Mvan_1534|M.vanbaalenii_PYR-1 RLVDAMAPRG--ARILDAGCGPGRVGGYLAAAGHQVVGVDVDPALIEAAE
MAB_2548c|M.abscessus_ATCC_199 RLVDALVPRG--ARILDAGCGAGRVGGFLAAAGHHVVGVDVDPALIEAAE
Mflv_3090|M.gilvum_PYR-GCK DLIEALLREGGGTRVLDAGCGTGRVAIELAARGFDVVGLDADPTMLETAR
Mb3374|M.bovis_AF2122/97 --IDWLLPAAA-RRVLDLGAGTGKLTTRLVERGLDVVAVDPIPEMLDVLR
Rv3342|M.tuberculosis_H37Rv --IDWLLPAAA-RRVLDLGAGTGKLTTRLVERGLDVVAVDPIPEMLDVLR
MLBr_01713|M.leprae_Br4923 VVYQWLAQRCTGCEVLEAGCGEGYGADLIVDVAYRVIAVDYDEATVAHVR
: : .:*: *.* * : . *:.:* : .
MSMEG_3430|M.smegmatis_MC2_155 EDYPGPRWLVGDLAELDLP-------------------ARGIAEPFDIIV
TH_1247|M.thermoresistible__bu QDYPGPQYLVGDLAELDLP-------------------ARGIPDPFDVIV
MMAR_1164|M.marinum_M QDYPGPQWLVGDLAELDLP-------------------ARGIAEPFDVIV
MUL_2851|M.ulcerans_Agy99 QDYPGPQWLVGDLAELDLP-------------------ARGIAEPFDVIV
MAV_4332|M.avium_104 HDHPGPRWLVGDLAELDLP-------------------ARGITDPFDVIV
Mvan_1534|M.vanbaalenii_PYR-1 QDYPGPRWLVGDLAELDLP-------------------ARGITEPFDLIV
MAB_2548c|M.abscessus_ATCC_199 HDHPGPRWLVGDLAELDLP-------------------SRGISEPFDVIV
Mflv_3090|M.gilvum_PYR-GCK AKAPRLRWIEADLVDTDD----------------------HLDETFDVVA
Mb3374|M.bovis_AF2122/97 AALPQTVALLGTAEEIPLDDNSVDAVL-VAQAWHWVDPARAIPEVARVLR
Rv3342|M.tuberculosis_H37Rv AALPQTVALLGTAEEIPLDDNSVDAVL-VAQAWHWVDPARAIPEVARVLR
MLBr_01713|M.leprae_Br4923 SRYPRVQVMQANLIELPLADASVDVVVNFQVIEHLWNQAQFVSECARVLR
* : . : : : ::
MSMEG_3430|M.smegmatis_MC2_155 SAGNV------MTFLAPSTRAQVLSRLRAHLTDNGRAVIGFGAGR-DYGF
TH_1247|M.thermoresistible__bu MAGNV------MAFLAPSTRTTVLSRLRAHLAEDGRAAIGFGAGR-DYDF
MMAR_1164|M.marinum_M SAGNV------MTFLAPSTRARVLGRLRAHLCSDGRAVIGFGAGR-DYAF
MUL_2851|M.ulcerans_Agy99 SAGNV------MTFLAPSTRARVLGRLRAHLCSDGRAVIGFGAGR-DYAF
MAV_4332|M.avium_104 SAGNV------MTFLAPSTRVLVLSRLRAHLAADGRAAIGFGAYR-EYEF
Mvan_1534|M.vanbaalenii_PYR-1 SAGNV------MTFLAPSTRVQVLARLRAHLRDDGRAVIGFGAGR-EYEY
MAB_2548c|M.abscessus_ATCC_199 SAGNV------MAFLAPSTRRQVLRRLRSHLADPGRAVIGFGAGR-DYEF
Mflv_3090|M.gilvum_PYR-GCK LPGNV------MIFLDRGTEAAVVAALARRLAPGGLLVAGFQLGTGRLTL
Mb3374|M.bovis_AF2122/97 PGGRLG-----LVWNTRDERLGWVRELGEIIGRDGDPVRDRVTLPEPFTT
Rv3342|M.tuberculosis_H37Rv PGGRLG-----LVWNTRDERLGWVRELGEIIGRDGDPVRDRVTLPEPFTT
MLBr_01713|M.leprae_Br4923 PSGLLMVSTPNRITFSPGRDTSINPFHTRELNADELTQLLITADFSEVAM
* : . :
MSMEG_3430|M.smegmatis_MC2_155 E--------EFLADAKGAGFTPDQLLSTWDVRPFSEDSDFLVAILRRA--
TH_1247|M.thermoresistible__bu D--------EFLADAATAGLTPDLLLSTWDLRPFTEDSEFLVALLRRA--
MMAR_1164|M.marinum_M A--------DFLDDAQAAGLVPDLLLSTWDLRPFTDDCDFLVALLRPA--
MUL_2851|M.ulcerans_Agy99 A--------DFLDDAQAAGLVPDLLLSTWDLRPFTDDCDFLVALLRPA--
MAV_4332|M.avium_104 T--------DFLNDAADAGFAPDLLLSSWDLRPFTEDSDFLVAVLRPA--
Mvan_1534|M.vanbaalenii_PYR-1 P--------QFLDDAAAGGLTPDLLLSTWDVRPFTDDSDFLVAVLRPA--
MAB_2548c|M.abscessus_ATCC_199 S--------EFLSDADAAGLVPDLLLSTWDLRPFTDKSDFLVALLRRDDA
Mflv_3090|M.gilvum_PYR-GCK D--------RYDEITATAALELVDRWATWDRQPY-DGGDYAVSLHRLT--
Mb3374|M.bovis_AF2122/97 V--------QRHQVEWTNYLTPQALIDLVASRSYCITSPAQVRTKTLDRV
Rv3342|M.tuberculosis_H37Rv V--------QRHQVEWTNYLTPQALIDLVASRSYCITSPAQVRTKTLDRV
MLBr_01713|M.leprae_Br4923 YGLFHGPRLKEMDARHGGSIIDAQIARAVSNAPWPPELVADVAAVTAADF
: .: *
MSMEG_3430|M.smegmatis_MC2_155 ------------------------------
TH_1247|M.thermoresistible__bu ------------------------------
MMAR_1164|M.marinum_M ------------------------------
MUL_2851|M.ulcerans_Agy99 ------------------------------
MAV_4332|M.avium_104 ------------------------------
Mvan_1534|M.vanbaalenii_PYR-1 ------------------------------
MAB_2548c|M.abscessus_ATCC_199 PS----------------------------
Mflv_3090|M.gilvum_PYR-GCK ------------------------------
Mb3374|M.bovis_AF2122/97 RQLLATHPALANSNGLALPYVTVCVRATLA
Rv3342|M.tuberculosis_H37Rv RQLLATHPALANSNGLALPYVTVCVRATLA
MLBr_01713|M.leprae_Br4923 DIIEASAGPEASDGRDVDESLDLIAIAVRR