For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MADAQLQLAALPAAEGRRAEQVMAAVRAALDAGVMRPGVKYSVYQLADALQVSRTPVRDALLRLEEIGLI RFEARQGFRILLPDPREIADIVAIRFALEPPAAGRAAAVCGPPLADRLAERMELLRRAAARGDEPGFAAH DLLLHDHILEAAGNNRARRIVRSLRESTRLLGGITADRSRTMSDIDAEHQPVVDAVIANDPAAAAAAMRT HLTSTGRLLVAQAVRDQDSPLDAAAVWAEAVGANTELP
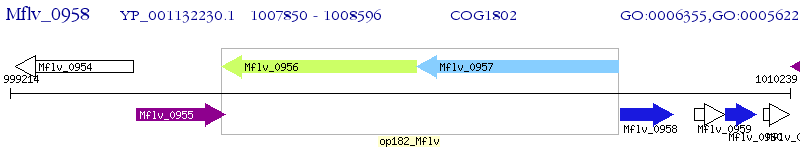
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0958 | - | - | 100% (248) | GntR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4061 | - | 1e-09 | 31.16% (215) | regulatory protein GntR, HTH |
| M. gilvum PYR-GCK | Mflv_4366 | - | 2e-08 | 25.71% (210) | GntR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0505 | - | 1e-10 | 27.85% (237) | GntR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0494 | - | 1e-10 | 27.85% (237) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4411c | - | 7e-17 | 29.63% (216) | GntR family transcriptional regulator |
| M. marinum M | MMAR_4029 | - | 1e-11 | 27.51% (229) | GntR family transcriptional regulator |
| M. avium 104 | MAV_0339 | - | 2e-10 | 25.73% (206) | transcriptional regulator, GntR family protein |
| M. smegmatis MC2 155 | MSMEG_4057 | - | 1e-18 | 31.53% (203) | transcriptional regulator, GntR family protein |
| M. thermoresistible (build 8) | TH_3342 | - | 1e-10 | 28.43% (204) | PUTATIVE transcriptional regulator, GntR family |
| M. ulcerans Agy99 | MUL_3894 | - | 1e-11 | 27.51% (229) | GntR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2942 | - | 6e-12 | 31.36% (220) | regulatory protein GntR, HTH |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0505|M.bovis_AF2122/97 MVEPMNQSSVFQPPDRQRVDERIATTIADAILDGVFPPGSTLPPERDLAE
Rv0494|M.tuberculosis_H37Rv MVEPMNQSSVFQPPDRQRVDERIATTIADAILDGVFPPGSTLPPERDLAE
MMAR_4029|M.marinum_M -------MTDPEFAARPQLSDDVARYVRKRIYEGNYAAGKYIRLD-QLAA
MUL_3894|M.ulcerans_Agy99 -------MTDPEFAARPQLSDDVARYVRKRIYEGNYAAGKYIRLD-QLAA
TH_3342|M.thermoresistible__bu -------VAVPEFAARPQLAEDVARYVRRRIFDGTYAAGEYVRLD-QLAV
MAV_0339|M.avium_104 -------MAAIDISG--TVRERAARELRDRILTGALPAGSRIDLD-AITA
Mvan_2942|M.vanbaalenii_PYR-1 --------MALRPVNRRSVPEDVFDQIISDVLSGEMRPGETLPSERRLAE
MSMEG_4057|M.smegmatis_MC2_155 ------------------MKDDVLAVLRDAIIAGKFAPGERLNEK-DLAE
Mflv_0958|M.gilvum_PYR-GCK -MADAQLQLAALPAAEGRRAEQVMAAVRAALDAGVMRPGVKYSVY-QLAD
MAB_4411c|M.abscessus_ATCC_199 --MTTLVPEEFAPLERQSTAELIAERLRIAIMRGVLAPGAQLGEA-SLAA
: : : * .* ::
Mb0505|M.bovis_AF2122/97 RLGVNRTSLRQGLARLQQMGLIEVRHGSGSVVRDPEGLTHPAVVEALVRK
Rv0494|M.tuberculosis_H37Rv RLGVNRTSLRQGLARLQQMGLIEVRHGSGSVVRDPEGLTHPAVVEALVRK
MMAR_4029|M.marinum_M ELGISVTPVREALFALRAEGLVAQQPHRGFVVLP----------------
MUL_3894|M.ulcerans_Agy99 ELGISVTPVREALFALRAEGLVAQQPHRGFVVLP----------------
TH_3342|M.thermoresistible__bu ELGVSVTPVREALFQLRAEGLLVQQPRRGFMVLP----------------
MAV_0339|M.avium_104 EFATSRTPVREALLELSFEGLVQVAPRSGVTVIG----------------
Mvan_2942|M.vanbaalenii_PYR-1 VLGVSRPAIREALKRLAAAGLIEVRQGDATTVRDFRRTAGLDLLPRLLIR
MSMEG_4057|M.smegmatis_MC2_155 QLGTSRGPIRDSLAALAHEGLVVHEPHKGARVPL----------------
Mflv_0958|M.gilvum_PYR-GCK ALQVSRTPVRDALLRLEEIGLIRFEARQGFRILLP---------------
MAB_4411c|M.abscessus_ATCC_199 QFAVSRGPLREAMQRLVSEGLLRSERNRGIFVIE----------------
: . .:*:.: * **: . :
Mb0505|M.bovis_AF2122/97 ---LGPDFLVELLEIRAALGPLIGRLAAARSTPEDAEALCAALEVVQQAD
Rv0494|M.tuberculosis_H37Rv ---LGPDFLVELLEIRAALGPLIGRLAAARSTPEDAEALCAALEVVQQAD
MMAR_4029|M.marinum_M ---LTKRDLADVAYVQAHVGGELAARAAINIGAAQLAELQQIQAQLEEAY
MUL_3894|M.ulcerans_Agy99 ---LTKRDLADVAYVQAHVGGELAARAAINIGAAQLAELQQIQAQLEEAY
TH_3342|M.thermoresistible__bu ---VTERDLTDIANVQAHVGGELAARAAQNITDEQLRVLTAIQDELEEAY
MAV_0339|M.avium_104 ---ISPEDVLDSFTILGVLTGQAAAWAAERIGPEELATLRELAAEVVAKS
Mvan_2942|M.vanbaalenii_PYR-1 DGDLDLAVVRSILETRLHNGPKVAELAAQRRGPELSDELNTAMRALADEA
MSMEG_4057|M.smegmatis_MC2_155 ---LDKADIEDVYTLRVALETLAARTAVERARAADFDALDHALSDLAKAF
Mflv_0958|M.gilvum_PYR-GCK ----DPREIADIVAIRFALEPPAAGRAAAVCGPPLADRLAERMELLRRAA
MAB_4411c|M.abscessus_ATCC_199 ---LTDDDVLDVYRSRSAIERAALECIMTGDTVTAYRRLLVPVAVMTEAA
: . * :
Mb0505|M.bovis_AF2122/97 T---AAARQAADLAYFRVLIHSTRNRALGLLYRWVEHAFGGREHALTGAY
Rv0494|M.tuberculosis_H37Rv T---AAARQAADLAYFRVLIHSTRNRALGLLYRWVEHAFGGREHALTGAY
MMAR_4029|M.marinum_M AADDDERTVRLNHEFHRAINLAADSPRLSQLMSQITRYQ--PESVFPGIE
MUL_3894|M.ulcerans_Agy99 AADDDERTVRLNHEFHRAINLAADSPRLSQLMSQITRYQ--PESVFPGIE
TH_3342|M.thermoresistible__bu TRDDEDRAVRLNHEFHRAINVAADSPKLAQVMSQITRYA--PESVFPTIS
MAV_0339|M.avium_104 G---DDSIGEANWHFHQMIHRAAHSPRLANQIKHAARVV--PTNFLTLFP
Mvan_2942|M.vanbaalenii_PYR-1 D---PVAAQRHALDFWDHIVDGADSIAFRLMFNTLRATYEPALPALATMM
MSMEG_4057|M.smegmatis_MC2_155 EKGDRRGITDADLRFHDAFYQAAHHDRLSVAWRTVRSQVALCLFSRNTVS
Mflv_0958|M.gilvum_PYR-GCK ARGDEPGFAAHDLLLHDHILEAAGNNRARRIVRSLRESTR-LLGGITADR
MAB_4411c|M.abscessus_ATCC_199 ARGDVVAVTDADQDFHEVLVDCSGSPRLRRAMRTLLLETRMLLGELQGAY
: :
Mb0505|M.bovis_AF2122/97 DDA-DPVLTDLRAINGAVLAGDPAAAAATVEAYLNASALRMVKSYRDRA-
Rv0494|M.tuberculosis_H37Rv DDA-DPVLTDLRAINGAVLAGDPAAAAATVEAYLNASALRMVKSYRDRA-
MMAR_4029|M.marinum_M HWP-ARAVKDHRRVLVALKKHDDKAARTAMSEHLASSAVPLIDHLIARGV
MUL_3894|M.ulcerans_Agy99 HWP-ARAVKDHGQVLVALKKHDDKAARTAMSEHLASSAVPLIDHLIARGV
TH_3342|M.thermoresistible__bu GWP-ARSTEHHRRVLAALADRDPDGARAAMSEHLAAGAGPLIEHLKEHGV
MAV_0339|M.avium_104 EHE-KHSLDEHAQLLDALGDKDVERARAIAERHVLDAGRSLAEWLEQRRD
Mvan_2942|M.vanbaalenii_PYR-1 AAE-VGRVDAYRAVAEAVLAGDPAAARDNAHRLLEPATTALLDALHTLEE
MSMEG_4057|M.smegmatis_MC2_155 ATSREIVVDEHVHLLDLLRTRSESALVDAVRAHIETAYQRLVASYD----
Mflv_0958|M.gilvum_PYR-GCK SRTMSDIDAEHQPVVDAVIANDPAAAAAAMRTHLTSTGRLLVAQAVRDQD
MAB_4411c|M.abscessus_ATCC_199 PDL-SEQVREHEVLCAAIGAGDAPAAYRLIDEHMHDAVTRLMARRDPGSV
: : . : :
Mb0505|M.bovis_AF2122/97 --------------------
Rv0494|M.tuberculosis_H37Rv --------------------
MMAR_4029|M.marinum_M VAESAPSGSAAERARGG---
MUL_3894|M.ulcerans_Agy99 VAESAPSGSAAERARGG---
TH_3342|M.thermoresistible__bu IAQ-----------------
MAV_0339|M.avium_104 GER-----------------
Mvan_2942|M.vanbaalenii_PYR-1 PR------------------
MSMEG_4057|M.smegmatis_MC2_155 --------------------
Mflv_0958|M.gilvum_PYR-GCK SPLDAAAVWAEAVGANTELP
MAB_4411c|M.abscessus_ATCC_199 E-------------------