For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IGRKDIRVPLSTARRSGLVEQVIDQLRHAVAGQEWRIGDRIPNETVLVEKFGVGRNTVREAVRALAHAGI LEVRQGDGTYVRATSEVSGALRRLCGEELREVLEVRRALEVEAARLAASRRTPADVAEMRGLLARRDERD SAGDVEAFARIDTEFHLAVVRCARNNTLIELYRGLLEAVTASVAHTRHVPVDVCDHGVLLDHIAAGDPDL AARTAGEFLDRIINGLGSADSS*
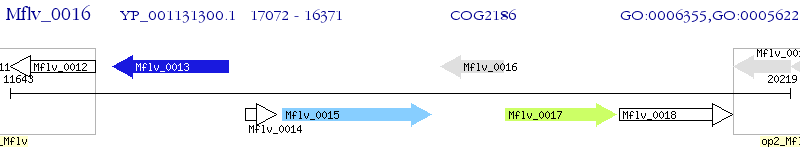
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0016 | - | - | 100% (233) | GntR domain-containing protein |
| M. gilvum PYR-GCK | Mflv_2114 | - | 3e-16 | 33.95% (215) | GntR domain-containing protein |
| M. gilvum PYR-GCK | Mflv_4061 | - | 3e-14 | 29.72% (212) | regulatory protein GntR, HTH |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0505 | - | 4e-15 | 29.03% (217) | GntR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0494 | - | 4e-15 | 29.03% (217) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2272c | - | 6e-30 | 38.94% (208) | putative transcriptional regulator, GntR |
| M. marinum M | MMAR_0749 | - | 3e-37 | 42.41% (224) | GntR family transcriptional regulator |
| M. avium 104 | MAV_2560 | - | 2e-14 | 26.75% (228) | regulatory protein GntR, HTH |
| M. smegmatis MC2 155 | MSMEG_2794 | - | 3e-68 | 62.50% (216) | transcriptional regulator, GntR family protein |
| M. thermoresistible (build 8) | TH_4115 | - | 3e-15 | 31.34% (217) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_1381 | - | 6e-37 | 43.18% (220) | GntR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0952 | - | 1e-94 | 79.02% (224) | GntR domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0016|M.gilvum_PYR-GCK MIGRKDIRVPLSTARRSGLVEQVIDQLRHAVAGQEWRIGDRIPNETVLVE
Mvan_0952|M.vanbaalenii_PYR-1 --------MPLSTARRTGLVDQVIEQLRRAVAGGEWRVGERIPNETVLVE
MSMEG_2794|M.smegmatis_MC2_155 --------MPLASTRRTGLVDQVIEQLRASVSSGEWPVGTKIPTEPELAD
MMAR_0749|M.marinum_M ----------MQTVRRRTLIAQVSDQLRDEITSGRWKVGDRIPTEPELCE
MUL_1381|M.ulcerans_Agy99 -------------MRRRTLIAQVSDQLRDEITSGRWKVGDRIPTEPELCE
MAB_2272c|M.abscessus_ATCC_199 ----------MQPVRRQTLIGQVTEQLREEIVSGRWAIGARIPTEPELCE
Mb0505|M.bovis_AF2122/97 MVEPMNQSSVFQPPDRQRVDERIATTIADAILDGVFPPGSTLPPERDLAE
Rv0494|M.tuberculosis_H37Rv MVEPMNQSSVFQPPDRQRVDERIATTIADAILDGVFPPGSTLPPERDLAE
TH_4115|M.thermoresistible__bu ---------------VQSSYIQVADQLRDLILRGDLAVGQKLPSEAEMAP
MAV_2560|M.avium_104 --------MEVTSLREPKMADRVATVLRRMFIRGEITEGTMLPPESELME
:: : . * :* * :
Mflv_0016|M.gilvum_PYR-GCK KFGVGRNTVREAVRALAHAGILEVRQ--GDGTYVRATSEVSG--------
Mvan_0952|M.vanbaalenii_PYR-1 SLGVGRNTVREAVRALAHAGILEVRQ--GDGTYVRATSEVSG--------
MSMEG_2794|M.smegmatis_MC2_155 TLGVGRNTLREAVRALAHSGILEVRQ--GDGTYVRATSEVSG--------
MMAR_0749|M.marinum_M MTGTARNTVREAVQALVHVGILERRQ--GSGTYVIADVEQGS--------
MUL_1381|M.ulcerans_Agy99 MTGTARNTVREAVQALVHVGILERRQ--GSGTYVIADVEQGS--------
MAB_2272c|M.abscessus_ATCC_199 LTGTSRNTIREAVQALVHAGMLERRQ--GSGTYVLS-LGGSS--------
Mb0505|M.bovis_AF2122/97 RLGVNRTSLRQGLARLQQMGLIEVRH--GSGSVVRDPEGLTHP---AVVE
Rv0494|M.tuberculosis_H37Rv RLGVNRTSLRQGLARLQQMGLIEVRH--GSGSVVRDPEGLTHP---AVVE
TH_4115|M.thermoresistible__bu LLGVSRSTIREALRILTTEHLIETRRGIQGGAFVAVPDSERLEGMFNTSF
MAV_2560|M.avium_104 RFGVSRPTLREAFRVLESESLIQVQRGVRGGARVTRPRRETLAR----YA
*. * ::*:.. * ::: :: .*: *
Mflv_0016|M.gilvum_PYR-GCK ALRR-LCGEELREVLEVRRALEVEAARLAASRRTPADVAEMRGLLARRD-
Mvan_0952|M.vanbaalenii_PYR-1 ALRR-LCGAELREVLQVRRALEVEGARLAAAHCTPADIAEMRTLLARRD-
MSMEG_2794|M.smegmatis_MC2_155 ALRR-LCGSELREVLQVRRCLEVEGARLAASNRTDADLAELRAFLDR---
MMAR_0749|M.marinum_M ALAEYFAAARERDLWELREAVEVTAAVLSARRRDAADIEDLRALLARRNE
MUL_1381|M.ulcerans_Agy99 ALAEYFAAARERDLWELREAVEVTAAVLSARRRDAADIEDLRVLLARRNE
MAB_2272c|M.abscessus_ATCC_199 AIADYFAAANNRDLIELRQTLEVTAAGLAAQRRDEDDIEQLRALLVRRNE
Mb0505|M.bovis_AF2122/97 ALVRKLGPDFLVELLEIRAALGPLIGRLAAARSTPEDAEALCAALEVVQ-
Rv0494|M.tuberculosis_H37Rv ALVRKLGPDFLVELLEIRAALGPLIGRLAAARSTPEDAEALCAALEVVQ-
TH_4115|M.thermoresistible__bu SMLALTNQVDARSFLQAMRALEGPAAALAAAARGGDQVEELKRTAAMVD-
MAV_2560|M.avium_104 GLILEYEGVTVKDVYDARVTLEVPMVEQLAKDRNPKVIAELEQIVERES-
.: .. : : * :
Mflv_0016|M.gilvum_PYR-GCK ----ERDSAGDVEAFARIDTEFHLAVVRCARNNTLIELYRGLLEAVT---
Mvan_0952|M.vanbaalenii_PYR-1 ----ECAATDDTDAFARTDAEFHLAVVRGSGNRTLTELYRGLIEVVT---
MSMEG_2794|M.smegmatis_MC2_155 ------TETSDDDDFVHSDTAFHLAVVRASHNSVLIELYRGLIEAIT---
MMAR_0749|M.marinum_M LWSREPANDKERTEMTSADTALHRAIVAASHNEIYLQFYDLLLPTIH---
MUL_1381|M.ulcerans_Agy99 LWSREPANDKERTEMTSADTALHRAIVAASHNETYLQFYDLLLPTIH---
MAB_2272c|M.abscessus_ATCC_199 LWAPHHVEPHNVEEAISTDIAVHRAVVAASHNALYLELYDSLLGLMD---
Mb0505|M.bovis_AF2122/97 -------QADTAAARQAADLAYFRVLIHSTRNRALGLLYRWVEHAFGGR-
Rv0494|M.tuberculosis_H37Rv -------QADTAAARQAADLAYFRVLIHSTRNRALGLLYRWVEHAFGGR-
TH_4115|M.thermoresistible__bu --------PADQEAAMQQSADFHSAILRASGNKLLEAMGRPVSAVARSK-
MAV_2560|M.avium_104 -------QLNPGSEAVDQLTDFHAAIARLSGNSTLQIVSDMLHHIIEKAN
. .: : * . :
Mflv_0016|M.gilvum_PYR-GCK ----ASVAHTRHVPVD--VCDHGVLLDHIAAGDPDLAARTAGEFLDRIIN
Mvan_0952|M.vanbaalenii_PYR-1 ----ASVATTRHVPVE--MVDHGVLVDQIAAGDAEQAARTAGELLDRILA
MSMEG_2794|M.smegmatis_MC2_155 ----ASVATASARPDG--KVSHRGLVEAIAAGDVERAGREAGGFLDDLLA
MMAR_0749|M.marinum_M ----RAVEAGPVGTCTSYEREHTELVEAIIAGDPQRAEAAAQVFIARLRP
MUL_1381|M.ulcerans_Agy99 ----RAVEAGPVGTCTSYEREHTELVEAIIAGDPQRAEAAAQVFIARLRR
MAB_2272c|M.abscessus_ATCC_199 ----HYMRGNPIGAECSFEHEHTRLVESVIAGDIGGATSATEQLFAELRL
Mb0505|M.bovis_AF2122/97 ----EHALTGAYDDADPVLTDLRAINGAVLAGDPAAAAATVEAYLNASAL
Rv0494|M.tuberculosis_H37Rv ----EHALTGAYDDADPVLTDLRAINGAVLAGDPAAAAATVEAYLNASAL
TH_4115|M.thermoresistible__bu ----FRRTVPGLEFWERNQRAHQELLKAILAGDENLAYELATRHIDEDLS
MAV_2560|M.avium_104 RSLQPTKGTRAEQAVRRSAKTHRMVLDMIKEGDAEKAGELWKRHLQKAEE
: : ** * :
Mflv_0016|M.gilvum_PYR-GCK GLGSADSS--------
Mvan_0952|M.vanbaalenii_PYR-1 GVDSADSC--------
MSMEG_2794|M.smegmatis_MC2_155 QTPDPQG---------
MMAR_0749|M.marinum_M ----------------
MUL_1381|M.ulcerans_Agy99 ----------------
MAB_2272c|M.abscessus_ATCC_199 SRDQPGRNFPHHPHD-
Mb0505|M.bovis_AF2122/97 RMVKSYRDRA------
Rv0494|M.tuberculosis_H37Rv RMVKSYRDRA------
TH_4115|M.thermoresistible__bu PYYLPPEEADRDGAGA
MAV_2560|M.avium_104 FVLTGAELSTVVDLLE