For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MALQPVTRRSVPEEVFEQIATDVLTGEMPPGEALPSERRLAELLGVSRPAVREALKRLSAAGLVEVRQGD VTTVRDFRRHAGLDLLPRLLFRNGELDISVVRSILEARLRNFPKVAELAAERNEPELAELLQDSLRALDT EEDPIVWQRHTLDFWDHVVDSAGSIVDRLMYNAFRAAYEPTLAALTTTMTAAAKRPSDYRKLADAICSGD PTGAKKAAQDLLELANTSLMAVLVSQASRQ
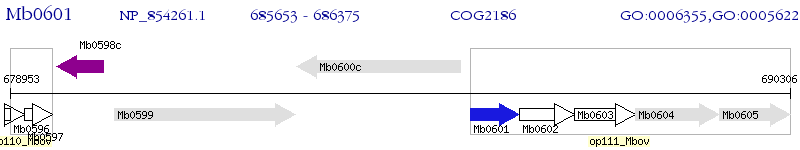
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0601 | - | - | 100% (240) | GntR family transcriptional regulator |
| M. bovis AF2122 / 97 | Mb0505 | - | 2e-21 | 31.80% (239) | GntR family transcriptional regulator |
| M. bovis AF2122 / 97 | Mb0816c | - | 2e-05 | 36.99% (73) | GntR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0016 | - | 7e-14 | 32.16% (227) | GntR domain-containing protein |
| M. tuberculosis H37Rv | Rv0586 | - | 1e-132 | 100.00% (240) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3018 | - | 4e-51 | 45.53% (235) | GntR family transcriptional regulator |
| M. marinum M | MMAR_0820 | - | 3e-22 | 32.73% (220) | GntR family transcriptional regulator |
| M. avium 104 | MAV_1821 | - | 1e-09 | 28.91% (211) | regulatory protein GntR, HTH |
| M. smegmatis MC2 155 | MSMEG_3527 | - | 2e-79 | 66.52% (233) | putative HTH-type transcriptional regulator |
| M. thermoresistible (build 8) | TH_1959 | - | 2e-85 | 68.67% (233) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (GNTR-FAMILY) |
| M. ulcerans Agy99 | MUL_4564 | - | 3e-22 | 32.73% (220) | GntR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2942 | - | 8e-83 | 66.09% (233) | regulatory protein GntR, HTH |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0601|M.bovis_AF2122/97 --------MALQPVTRRSVPEEVFEQIATDVLTGEMPPGEALPSERRLAE
Rv0586|M.tuberculosis_H37Rv --------MALQPVTRRSVPEEVFEQIATDVLTGEMPPGEALPSERRLAE
TH_1959|M.thermoresistible__bu --------MALQPVNRRSVPEDVFDQIVGEVLSGQMQPGDALPSERRLAE
Mvan_2942|M.vanbaalenii_PYR-1 --------MALRPVNRRSVPEDVFDQIISDVLSGEMRPGETLPSERRLAE
MSMEG_3527|M.smegmatis_MC2_155 -----MAVMALQPVNRRSVPEDVFEQIVSEVLTGEMRPGESLPSERRLAE
MAB_3018|M.abscessus_ATCC_1997 --------MALQPIARQSIPDEIFSQLAAQVLTGDRIPGDTLPSERALAE
MMAR_0820|M.marinum_M ----MNQSSILQPPDRQRVDEQIATSIADAILDGVFPPGSALPPERELAE
MUL_4564|M.ulcerans_Agy99 ----MNQSSILQPPDRQRVDEQIATSIADAILDGVFPPGSALPPERELAE
Mflv_0016|M.gilvum_PYR-GCK MIGRKDIRVPLSTARRSGLVEQVIDQLRHAVAGQEWRIGDRIPNETVLVE
MAV_1821|M.avium_104 --------MEVTSLREPKMADRVATVLRRMFIRGEIPEGTMLPPESELME
: . . : : : : . * :* * * *
Mb0601|M.bovis_AF2122/97 LLGVSRPAVREALKRLSAAGLVEVRQGD--VTTVRDFRRHAGLDLLPRLL
Rv0586|M.tuberculosis_H37Rv LLGVSRPAVREALKRLSAAGLVEVRQGD--VTTVRDFRRHAGLDLLPRLL
TH_1959|M.thermoresistible__bu VLGVSRPAVREALKRLSAAGLVEVRQGE--ITTVRDYRRHAGLDLLPRLL
Mvan_2942|M.vanbaalenii_PYR-1 VLGVSRPAIREALKRLAAAGLIEVRQGD--ATTVRDFRRTAGLDLLPRLL
MSMEG_3527|M.smegmatis_MC2_155 VLGVSRPAVREAIKRLTAAGLVEVRQGD--ATTVRDFRRHAGMDLLPRLL
MAB_3018|M.abscessus_ATCC_1997 ALGVSRAAVREALGRLERSGLIQIRQGG--STVIRDYRSDAGFDVLPLLL
MMAR_0820|M.marinum_M QLGVNRTSLRQGLARLQQMGLIETRQGS--GNVVRDPQSLTHPAVVEALV
MUL_4564|M.ulcerans_Agy99 QLGVNRTSLRQGLARLQQMGLIETRQGS--GNVVRDPQSLTHPAVVEALV
Mflv_0016|M.gilvum_PYR-GCK KFGVGRNTVREAVRALAHAGILEVRQGD--GTYVR-----ATSEVSGALR
MAV_1821|M.avium_104 RFGVSRPTLREAFRVLESESLIVVQRGVRGGARVTRPRRETLARYAGLIL
:**.* ::*:.. * .:: ::* : : :
Mb0601|M.bovis_AF2122/97 FRNGELDISVVRSILEARLRNF-PKVAELAAER--NEPELAELLQDSLRA
Rv0586|M.tuberculosis_H37Rv FRNGELDISVVRSILEARLRNF-PKVAELAAER--NEPELAELLQDSLRA
TH_1959|M.thermoresistible__bu FRAGELDVTVVRSILETRLHNG-PKVAELAARR--RPPGLGALLDEAVQT
Mvan_2942|M.vanbaalenii_PYR-1 IRDGDLDLAVVRSILETRLHNG-PKVAELAAQR--RGPELSDELNTAMRA
MSMEG_3527|M.smegmatis_MC2_155 FRAGELDTSVVRSILETRLHNG-PKVAELAAER--RGAELVGQLTAVVHA
MAB_3018|M.abscessus_ATCC_1997 AYGGDVDRRTLASVIEARAIIG-PQVARLAAQRAHQTPGVADRLRAMTSE
MMAR_0820|M.marinum_M RKLG---PDFLVEVLEIRAALG-PLIGRLAAER--NSSEDAEALRAALAE
MUL_4564|M.ulcerans_Agy99 RKLG---PDFLVEVLEIRAALG-PLIGRLAAER--NSSEDAEALRAALAE
Mflv_0016|M.gilvum_PYR-GCK RLCG----EELREVLEVRRALE-VEAARLAASR--RTPADVAEMRGLLAR
MAV_1821|M.avium_104 EYEG----VTLKDVYDARVTLETPMIVQLAKDR---NPAVIAELEQIVER
* : .: : * .** * . :
Mb0601|M.bovis_AF2122/97 LDTEEDPIVWQRHTLDFWDHVVDSAGSIVDRLMYNAFRAAYEPTLAALT-
Rv0586|M.tuberculosis_H37Rv LDTEEDPIVWQRHTLDFWDHVVDSAGSIVDRLMYNAFRAAYEPTLAALT-
TH_1959|M.thermoresistible__bu LAAETDPVERQRHALRFWDHVVDGADSIAFRLMYNTLRATYEPALPALA-
Mvan_2942|M.vanbaalenii_PYR-1 LADEADPVAAQRHALDFWDHIVDGADSIAFRLMFNTLRATYEPALPALA-
MSMEG_3527|M.smegmatis_MC2_155 LEAEIDPIQRQRHAMTFWDHVVDGADSIAFRLMFNTLRETYEPVLPALA-
MAB_3018|M.abscessus_ATCC_1997 LEAEDDPLQRMWQALGFWELVIDTADSIAFRLVFNTMRDSYVRALDVLV-
MMAR_0820|M.marinum_M VADADTAPARQAADLAYFRVLIHGTRNRALGLLYRWVEQAFGGREHELT-
MUL_4564|M.ulcerans_Agy99 VADADTAPARQAADLAYFRVLIHGTRNRALGLLYRWVEQAFGGREHDLT-
Mflv_0016|M.gilvum_PYR-GCK RDERDSAGDVEAFARIDTEFHLAVVRCARNNTLIELYRGLLEAVTASVA-
MAV_1821|M.avium_104 EAELKPGSDTVDQLTDFHAAIARLSGSKTLQIVSDMLHHIVEKANRSLQP
: . :
Mb0601|M.bovis_AF2122/97 ----TTMTAAAKRPSDYRKLADAICSGDPTGAKKAAQDLLELANTSLMAV
Rv0586|M.tuberculosis_H37Rv ----TTMTAAAKRPSDYRKLADAICSGDPTGAKKAAQDLLELANTSLMAV
TH_1959|M.thermoresistible__bu ----TMMAAEVGRPQAYRALAAAIEAGDPTAAEQSARDLLEPATAALLSA
Mvan_2942|M.vanbaalenii_PYR-1 ----TMMAAEVGRVDAYRAVAEAVLAGDPAAARDNAHRLLEPATTALLDA
MSMEG_3527|M.smegmatis_MC2_155 ----AVMAGEVADPDAYRAITRAISDRDADAARGAAQNLLEPATHALLGA
MAB_3018|M.abscessus_ATCC_1997 ----NVMAAEVGDIGHYRALADAIALADPDAAQDAAAGMLALGTKAFDKL
MMAR_0820|M.marinum_M ----GAYEDADPVIADLTAINDAVLAGDPETAAAAVEGYLNASALRMIMC
MUL_4564|M.ulcerans_Agy99 ----GAYEDADPVIADLTAINDAVLAGDPETAAAAVEGYLNASALRMIMC
Mflv_0016|M.gilvum_PYR-GCK ----HTRHVPVD-VCDHGVLLDHIAAGDPDLAARTAGEFLDRIINGLGSA
MAV_1821|M.avium_104 TEGTRAAQAVSRSAKTHRMVLDMIKAGEAEKAGELWKKHLQKAEAFVLSG
: : :. * * .
Mb0601|M.bovis_AF2122/97 LVSQASRQ----
Rv0586|M.tuberculosis_H37Rv LVSQASRQ----
TH_1959|M.thermoresistible__bu LDELEAQK----
Mvan_2942|M.vanbaalenii_PYR-1 LHTLEEPR----
MSMEG_3527|M.smegmatis_MC2_155 LDALEAAR----
MAB_3018|M.abscessus_ATCC_1997 LREMEKER----
MMAR_0820|M.marinum_M YRAAYAGEDPER
MUL_4564|M.ulcerans_Agy99 YRAAYAGEDPER
Mflv_0016|M.gilvum_PYR-GCK DSS---------
MAV_1821|M.avium_104 AELSTVVDLLE-