For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
CTVTATRVEVMEWPHELTPGDMAARSGVAVSALHFYEREGLITSRRTGGNQRRYARETLRRVAFIRMSQR LGIPLARIREALATLPSDRVPTSKDWARLSAGWRDDLDQRILHLERLRDNLAGCIGCGCLSLKICALANP GDVLADEGPGAARL*
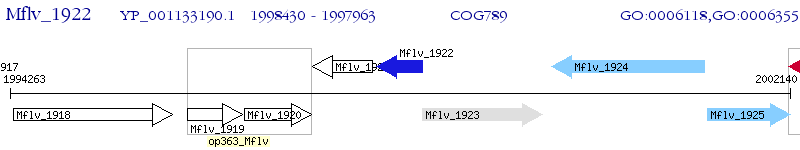
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1922 | - | - | 100% (155) | MerR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3212 | - | 2e-09 | 35.58% (104) | MerR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3401 | - | 1e-05 | 36.92% (65) | MerR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3367 | - | 2e-05 | 34.38% (64) | MerR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3334 | - | 2e-05 | 34.38% (64) | MerR family transcriptional regulator |
| M. leprae Br4923 | MLBr_02075 | - | 3e-05 | 34.85% (66) | hypothetical protein MLBr_02075 |
| M. abscessus ATCC 19977 | MAB_2309c | - | 4e-34 | 46.21% (145) | putative transcriptional regulator |
| M. marinum M | MMAR_2990 | hspR_2 | 2e-06 | 41.67% (60) | heat shock protein transcriptional repressor HspR_2 |
| M. avium 104 | MAV_3222 | - | 1e-09 | 32.77% (119) | transcriptional regulator, MerR family protein |
| M. smegmatis MC2 155 | MSMEG_5450 | soxR | 7e-70 | 87.86% (140) | redox-sensitive transcriptional activator SoxR |
| M. thermoresistible (build 8) | TH_0931 | soxR | 6e-68 | 87.86% (140) | redox-sensitive transcriptional activator SoxR |
| M. ulcerans Agy99 | MUL_0011 | - | 9e-06 | 38.81% (67) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_4811 | - | 9e-73 | 88.89% (144) | MerR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3367|M.bovis_AF2122/97 ------------------MKISEVAALTNTSTKTLRFYENSGLLPPPART
Rv3334|M.tuberculosis_H37Rv ------------------MKISEVAALTNTSTKTLRFYENSGLLPPPART
Mflv_1922|M.gilvum_PYR-GCK MCTVTATRVEVMEWPHELTP-GDMAARSGVAVSALHFYEREGLITS-RRT
Mvan_4811|M.vanbaalenii_PYR-1 -----------MESAHELTP-GEMAVRSGVAVSALHFYEREGLISS-RRT
MSMEG_5450|M.smegmatis_MC2_155 --------------MHELTP-GELAQRSGVAVSALHFYEREGLITS-RRT
TH_0931|M.thermoresistible__bu ------------MGAHELTP-SEMSARSGVAVSALHFYEREGLITS-RRT
MAB_2309c|M.abscessus_ATCC_199 -----------MKGKTELLPIGEVARRSGIAVSAVRYYADIGLVPA-ERT
MAV_3222|M.avium_104 -----------------MLTIGEVARRSGVAATTLRYYEQIGLLPAPTRL
MUL_0011|M.ulcerans_Agy99 ---------------MDGLTVGQVAERFGITVRTLHHYDEIGLLIPSRRA
MMAR_2990|M.marinum_M -MADRPGHAGGPAPDVGVYGISVTAELSGIAVQSLRLYERHGLVTP-ARS
MLBr_02075|M.leprae_Br4923 --MSAPDSPALVGMSIGAVLDLLRSDFPDVTISKIRFLEAEGLVTP-QRS
: . : :: **: . *
Mb3367|M.bovis_AF2122/97 ASGYRNYGPEIVDRLRFIHRGQAAG-LALQEVRQILAI------------
Rv3334|M.tuberculosis_H37Rv ASGYRNYGPEIVDRLRFIHRGQAAG-LALQEVRQILAI------------
Mflv_1922|M.gilvum_PYR-GCK GGNQRRYARETLRRVAFIRMSQRLG-IPLARIREALAT------------
Mvan_4811|M.vanbaalenii_PYR-1 SGNQRRYARDTLRRVAFIRMSQRLG-IPLARIREALAT------------
MSMEG_5450|M.smegmatis_MC2_155 SGNQRRYARETLRRVAFIRMSQRLG-IPLSRIREALST------------
TH_0931|M.thermoresistible__bu AGNQRRYPREMLRRVAFIRMSQRLG-IPLARIREALAT------------
MAB_2309c|M.abscessus_ATCC_199 SGNVRVFRRHALRRISLIRVATGFG-IPLSEVAEVLST------------
MAV_3222|M.avium_104 GG-QRRYDEAVLSRLEVIALCKTAG-FALDEIQRLFA-------------
MUL_0011|M.ulcerans_Agy99 ASGYRVYTSADLTRLSQVIVYRRLE-LPLDEIARLLDEGNAVSHLVRQRE
MMAR_2990|M.marinum_M EGGTRRYSADDLVRLQRVSVLIEAG-INLAGISRILNL------------
MLBr_02075|M.leprae_Br4923 ASGYRRFTAYDCARLRFILTAQRDHYLPLKVIRAQLDAQPDGELPSFG--
. * : *: : : * : :
Mb3367|M.bovis_AF2122/97 -----------------------------------------HDRGEAPCA
Rv3334|M.tuberculosis_H37Rv -----------------------------------------HDRGEAPCA
Mflv_1922|M.gilvum_PYR-GCK -----------------------------------------LPSDRVPTS
Mvan_4811|M.vanbaalenii_PYR-1 -----------------------------------------LPTDRVPTS
MSMEG_5450|M.smegmatis_MC2_155 -----------------------------------------LPNDRVPTS
TH_0931|M.thermoresistible__bu -----------------------------------------LPSDRVPTS
MAB_2309c|M.abscessus_ATCC_199 -----------------------------------------LPDYRAPST
MAV_3222|M.avium_104 --------------------------------------------DDAPGR
MUL_0011|M.ulcerans_Agy99 RVMARLDEMKDLVEAIDLALEKAMTNTPMTDDDMRELFGDGFDDYRVETE
MMAR_2990|M.marinum_M -------------------------------------------------E
MLBr_02075|M.leprae_Br4923 ----------------------------SPYVAPRLFSVTGGPGAGVGSG
Mb3367|M.bovis_AF2122/97 HVRQLLST-RIDEVRAQ---IAELIALEGHLQTLLDHASYGPPTEHDHST
Rv3334|M.tuberculosis_H37Rv HVRQLLST-RIDEVRAQ---IAELIALEGHLQTLLDHASYGPPTEHDHST
Mflv_1922|M.gilvum_PYR-GCK KDWARLSAGWRDDLDQR---ILHLERLRDNLAGCIGCGCLSLKICALANP
Mvan_4811|M.vanbaalenii_PYR-1 KDWARLSAGWREDLDQR---IVHLQRLRDNLAGCIGCGCLSLKVCALANP
MSMEG_5450|M.smegmatis_MC2_155 KDWARLSAGWRQDLDDR---IMHLQRLRDNLTGCIGCGCLSLKTCQLTNP
TH_0931|M.thermoresistible__bu KDWAELSAGWRADLDER---ILHLQRLRDNLADCIGCGCLSLKTCALSNP
MAB_2309c|M.abscessus_ATCC_199 RDWQRISRRWHAHLEAR---KNAIADMQEKLTGCIGCGCLSMTKCALFNP
MAV_3222|M.avium_104 PASRALARAKLAQIDAQ---LDSLTRARAVIEWGMRCTCPSIDACTCGIH
MUL_0011|M.ulcerans_Agy99 QKWGETAEWKESQRRTKSYGKDEWVRIKAEGQAIEKALSDAFRAGLPADS
MMAR_2990|M.marinum_M DDNAALAAANCDLLASN-------RSLRDAARAAVRLMNHGRSAENGEGG
MLBr_02075|M.leprae_Br4923 VGSDIPAVSPAGVRLSREDLLDRSGVADDLLTALLKAGVITTGPGGFFDE
: .
Mb3367|M.bovis_AF2122/97 VCWILESDLDEPTAIEVSDIHA----------------------------
Rv3334|M.tuberculosis_H37Rv VCWILESDLDEPTAIEVSDIHA----------------------------
Mflv_1922|M.gilvum_PYR-GCK GDVLAD---EGPGAARL---------------------------------
Mvan_4811|M.vanbaalenii_PYR-1 GDVLSE---EGPGPVRL---------------------------------
MSMEG_5450|M.smegmatis_MC2_155 GDVLAE---RGPGAARL---------------------------------
TH_0931|M.thermoresistible__bu DDALAA---RGPGAARL---------------------------------
MAB_2309c|M.abscessus_ATCC_199 ADILAD---DGSTGALLLADLG----------------------------
MAV_3222|M.avium_104 PTRPAPVGPDVRSARRF---------------------------------
MUL_0011|M.ulcerans_Agy99 EEAMNAAEQHRRHVNRWFYDCPPAFHRNLGDMYVSDPRYIATYDESFGLP
MMAR_2990|M.marinum_M DTS-----------------------------------------------
MLBr_02075|M.leprae_Br4923 HAIVILQCARALSEYGVEPRHLRAFRSAADRQSDLIVQIAGPIVKAGKAG
Mb3367|M.bovis_AF2122/97 ----------------------------------
Rv3334|M.tuberculosis_H37Rv ----------------------------------
Mflv_1922|M.gilvum_PYR-GCK ----------------------------------
Mvan_4811|M.vanbaalenii_PYR-1 ----------------------------------
MSMEG_5450|M.smegmatis_MC2_155 ----------------------------------
TH_0931|M.thermoresistible__bu ----------------------------------
MAB_2309c|M.abscessus_ATCC_199 ----------------------------------
MAV_3222|M.avium_104 ----------------------------------
MUL_0011|M.ulcerans_Agy99 GLAAYCREAIHANANRAGG---------------
MMAR_2990|M.marinum_M ----------------------------------
MLBr_02075|M.leprae_Br4923 ARDRADDLAREVAALAITLHTSLIKSAVRGVLHR