For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DEGTAPHPDRDGLLDVVVDMLETDGYDAVQLRVVARRARSSLATIYKRYPTRDDLILAALEYWVARHRYA GIAVPPAAGLSLRQGLMSLFRTIFEPWERHPEMLKTYFRVRSSPNGGKLFRFGFDIVAPAGMELLKGVDE EFVVSLEAILANVIYGLLGRFSSGEIAVTEILPVLDRTVYWLTRGYEAPDSDSAEHFPTTPCAPTGR*
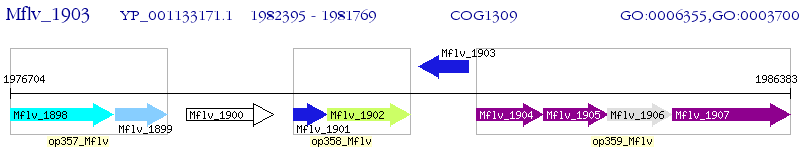
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1903 | - | - | 100% (208) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3021 | - | 1e-09 | 28.74% (174) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_4581 | - | 2e-06 | 23.43% (175) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1581 | - | 1e-08 | 33.67% (98) | regulatory protein |
| M. tuberculosis H37Rv | Rv1556 | - | 1e-08 | 33.67% (98) | regulatory protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0989 | - | 2e-06 | 32.46% (114) | putative HTH-type transcriptional regulator TetR |
| M. marinum M | MMAR_0929 | - | 2e-63 | 60.96% (187) | TetR family transcriptional regulator |
| M. avium 104 | MAV_3548 | - | 3e-52 | 52.58% (194) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_5811 | - | 1e-68 | 69.83% (179) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_2255 | - | 7e-07 | 42.86% (56) | TetR-family protein transcriptional regulator |
| M. ulcerans Agy99 | MUL_0681 | - | 8e-62 | 58.82% (187) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_3393 | - | 8e-60 | 61.02% (177) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1903|M.gilvum_PYR-GCK --------------MDEGTAPHPDRDGLLDVVVDMLETDGYDAVQLRVVA
MSMEG_5811|M.smegmatis_MC2_155 --------------MGEDIEERDDR--LLDVVTEILETEGYEAVQLREVA
MMAR_0929|M.marinum_M MTARVGTQLASPETVTAPAVRRDDR--ILDIVVHLLQTEGYDAVQLREVA
MUL_0681|M.ulcerans_Agy99 MTARVGTQLASPETVTAPAVRRDDR--ILDIVVHLLQTEGYDAVQLREVA
Mvan_3393|M.vanbaalenii_PYR-1 -------------------MGAEDP--ILTVVVDLLENEGYDAVQLREVA
MAV_3548|M.avium_104 ----------MTAKGPDAPIVSGDP--ILGIVVDMLDTGGYEAVQLREVA
Mb1581|M.bovis_AF2122/97 MVGAVTQ---IADRPTDPSPWSPRETELLAVTLRLLQEHGYDRLTVDAVA
Rv1556|M.tuberculosis_H37Rv MVGAVTQ---IADRPTDPSPWSPRETELLAVTLRLLQEHGYDRLTVDAVA
MAB_0989|M.abscessus_ATCC_1997 MASDVSTGLTKAARGRQVSQWTPREVELLKVTLRLLQEHGYDRLSVDEVA
TH_2255|M.thermoresistible__bu ----------------MPERPDPARQAILDAAVVEFEQHGFRKVALEDVA
:* . :: *: : : **
Mflv_1903|M.gilvum_PYR-GCK RRARSSLATIYKRYPTRDDLILAALEYWVARHRYAGI-AVPP-AAGLSLR
MSMEG_5811|M.smegmatis_MC2_155 RRARTSLATIYKRFATRDDLILAALEYWMQKNRYSGV-TPRRRTPGVSLY
MMAR_0929|M.marinum_M RRARTSLATIYKRYANRDELILAALEFWMDEHHYAGL-AEQTPAPGESLY
MUL_0681|M.ulcerans_Agy99 RRARTSLATIYKRYANRDELILAALEFWMDEHHYAGL-AEQTPAPGECLY
Mvan_3393|M.vanbaalenii_PYR-1 RRSRTSLTTIYKRYSNRDELILAALEAWTDTHRYAAV-AEQRREPGETLH
MAV_3548|M.avium_104 RRARVSMATIYKRYRTRDELIVAALEGWMDANRYARLPSLIDELPGKSMY
Mb1581|M.bovis_AF2122/97 ASARASKATVYRRWPSKAELVLAAFIEGIRQVAVPPN--------TGNLR
Rv1556|M.tuberculosis_H37Rv ASARASKATVYRRWPSKAELVLAAFIEGIRQVAVPPN--------TGNLR
MAB_0989|M.abscessus_ATCC_1997 AESKASKATIYRRWPSKAELVLAAFIEGMRAQLVRPN--------TGSLR
TH_2255|M.thermoresistible__bu RRAGVSRTTIYRRFTNRDELVAAVVDRENAALFADIAAELKHSGPQGDYY
: * :*:*:*: .: :*: *..
Mflv_1903|M.gilvum_PYR-GCK QGLMSLFRTIFEPWERHPEMLKTYFRVRSSPNGGKLFRFGFDIVAPAGME
MSMEG_5811|M.smegmatis_MC2_155 AALTGLLRTIFEPWERHPAMLEAYFRVRSSRSGQKLFRFGFDIVAPAGLE
MMAR_0929|M.marinum_M AGMMRVLRTIFQPWETHPDIVKAYFRARAAPGGQRLVHRGLDMVVPAAME
MUL_0681|M.ulcerans_Agy99 AGMMRVLRMIFQPWETHPDIVKAYFRARAAPGGQRLVHRGLDMVVPAAME
Mvan_3393|M.vanbaalenii_PYR-1 DALMRLFRVLFEPWERHPAMLAAYFRARTGPSGKRLLRRGLDVVVPAALD
MAV_3548|M.avium_104 SDLMHVMRTIFEPWERHPLMLRSYFQARSGPGGKRLIQRGVEAVVPVAKS
Mb1581|M.bovis_AF2122/97 DDLLRLGELICREVGQHASTIRAVLVEVSR---NPALNDVLQHQFVDHRK
Rv1556|M.tuberculosis_H37Rv DDLLRLGELICREVGQHASTIRAVLVEVSR---NPALNDVLQHQFVDHRK
MAB_0989|M.abscessus_ATCC_1997 EDLLRIGTNALADVRRNTPVMRGLLIEIER---NPALTKAFRTKFVDERK
TH_2255|M.thermoresistible__bu VEAFTLAILKFRRHR----VLHRMITDEPGFILHLAQQHWGAAVERMAEA
: : :
Mflv_1903|M.gilvum_PYR-GCK LLKGVDEEFVVSLEAILANVIYGLLGRFSSGEIAVTEILPVLDRTVYWLT
MSMEG_5811|M.smegmatis_MC2_155 LLADVDDEFVADLDTIVSTVVYGLLGRFAAGEIAVTDIVPILDRTVYWLT
MMAR_0929|M.marinum_M VLAGVDENFIHDLDTVISSLVYGLLGRFTAGEIAITEILPSIDRTVFWLI
MUL_0681|M.ulcerans_Agy99 VLAGVEEDFIHDLDTVISSLVYGLVGRFTAGEIAITEILPSIDRTVFWLI
Mvan_3393|M.vanbaalenii_PYR-1 VLAGVDEEFIADMDTVIANVIYGLLGRFAAGEITITDILPAIDRTVYRLT
MAV_3548|M.avium_104 VLAQADPGFVNDLELILTGVVFGFLTRFAQGEIEVTEIVPGIERTVYWLT
Mb1581|M.bovis_AF2122/97 ALIQYILQQAVDRGEISSAAISDELWDLLPGYLIFRSIIPNRPPTQDTVQ
Rv1556|M.tuberculosis_H37Rv ALIQYILQQAVDRGEISSAAISDELWDLLPGYLIFRSIIPNRPPTQDTVQ
MAB_0989|M.abscessus_ATCC_1997 ALIDDVLFAAVQRGEIDADAVNEELWDLLPGYLVFRSLLPVRSPTADTVR
TH_2255|M.thermoresistible__bu LRVIFPAGFAERIGEAAVIEFADTILRYAAMALLLPSVQPLDTADDIRAF
. : : . .: *
Mflv_1903|M.gilvum_PYR-GCK RGY---EAP-DSDSAEHFPTTPCAPTGR------
MSMEG_5811|M.smegmatis_MC2_155 TGY---EAAGNARPGERRRRSPARRHRN------
MMAR_0929|M.marinum_M RGY---EAS---RRAG------------------
MUL_0681|M.ulcerans_Agy99 RGY---EAS---RRAG------------------
Mvan_3393|M.vanbaalenii_PYR-1 AGYA-LEAAGDGVRRG------------------
MAV_3548|M.avium_104 QAYQNADAGRQPHISGGGISQPSEGSTSSLYCGQ
Mb1581|M.bovis_AF2122/97 ALVDDVILPSLTRSTG------------------
Rv1556|M.tuberculosis_H37Rv ALVDDVILPSLTRSTG------------------
MAB_0989|M.abscessus_ATCC_1997 TLVDDVIMPSLTRER-------------------
TH_2255|M.thermoresistible__bu ATRHFLPSLPAALHTGSA----------------