For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NLAGRVALVTGGASGIGRATAVRLAAEGMQVCVVDIDGSAAEEVAESFGGIGLRCDVTDPDQVDATFAAC AAGFGRLDLAFLNAGVTIDWSGDIGSLDVAQYRRSVGVNVDGVVFGVRAAVRTMRSLGAGTDRAILATAS LAGLMPWHPDPVYSLGKHGVVGFMRSVAPNLAAESIAVHTICPGITETGVLGDRRPLVERIGVPVMEPDA IAEAVMVALDAPADATGTCWVAQYGKPAWPMVFAPVESPDSRLNVPVRLR*
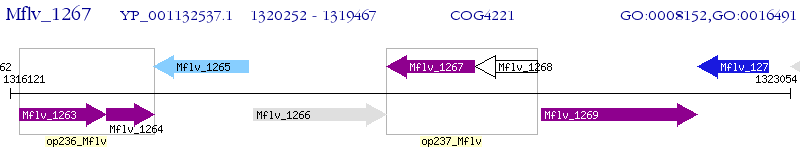
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1267 | - | - | 100% (261) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_3370 | - | 6e-25 | 35.90% (195) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_4369 | fabG | 5e-24 | 32.22% (239) | 3-ketoacyl-(acyl-carrier-protein) reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2882c | - | 3e-28 | 34.29% (245) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv2857c | - | 4e-28 | 34.29% (245) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 4e-18 | 33.70% (184) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_4335 | - | 4e-26 | 37.05% (224) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_4765 | - | 8e-27 | 38.54% (192) | short chain dehydrogenase |
| M. avium 104 | MAV_0886 | - | 8e-27 | 39.38% (193) | short-chain dehydrogenase/reductase SDR |
| M. smegmatis MC2 155 | MSMEG_6623 | - | 1e-110 | 76.06% (259) | dehydrogenase |
| M. thermoresistible (build 8) | TH_1621 | - | 1e-107 | 72.20% (259) | dehydrogenase |
| M. ulcerans Agy99 | MUL_0336 | - | 5e-27 | 38.54% (192) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_3101 | - | 4e-27 | 36.92% (195) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1267|M.gilvum_PYR-GCK ----MNLAG---------------RVALVTGGASGIGRATAVRLAAEGMQ
MSMEG_6623|M.smegmatis_MC2_155 ----MELQD---------------RVALVTGGGSGIGRAAAQRLAAHGMR
TH_1621|M.thermoresistible__bu ----VRLDG---------------RAALVTGGGSGIGKATAQRLAAAGMR
Mb2882c|M.bovis_AF2122/97 ---MMDLSQRLAG-----------RVAVITGGGSGIGLAAGRRMRAEGAT
Rv2857c|M.tuberculosis_H37Rv ---MMDLSQRLAG-----------RVAVITGGGSGIGLAAGRRMRAEGAT
MLBr_01807|M.leprae_Br4923 ---MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHR
MMAR_4765|M.marinum_M --MVDELAG---------------KIAIVTGGASGIGRATVARFIAEGAR
MUL_0336|M.ulcerans_Agy99 --MVDELAG---------------KIAIVTGGASGIGRATVARFIAEGAR
MAV_0886|M.avium_104 --MVNELAG---------------KVAIVTGGASGIGRGIVERFVAEGAR
MAB_4335|M.abscessus_ATCC_1997 MGTAVQLAG---------------KVVAVTGGARGIGRAIATAFAAEGAK
Mvan_3101|M.vanbaalenii_PYR-1 ---MAQLSG---------------KVVFITGGASGIGAALTARLGAAGAQ
: :*** *** . : * *
Mflv_1267|M.gilvum_PYR-GCK VCVVDIDGSAAE--EVAESFGGI---GLRCDVTDPDQVDATFAACAAGFG
MSMEG_6623|M.smegmatis_MC2_155 VCVVDIDGDAAR--EVAEPLGGL---ALTCDVSDPDQVNVTFDRCATELG
TH_1621|M.thermoresistible__bu VCVADIDADAAA--EVADTVNGV---AVRCDVSDPAQVDRVFAACTHELG
Mb2882c|M.bovis_AF2122/97 IVVGDVDVEAGG--AAADELSGL---FVPTDVCDEDAVNGLFDGAAETYG
Rv2857c|M.tuberculosis_H37Rv IVVGDVDVEAGG--AAADELSGL---FVPTDVCDEDAVNGLFDGAAETYG
MLBr_01807|M.leprae_Br4923 VAV----THRAS--VVPDWFFG-----VECDVTDNDAIGAAFKAVEEHQG
MMAR_4765|M.marinum_M VVIADVEEERGESLAAALGADAM---FCRTDVSQPEQVAAVVAAAVENFG
MUL_0336|M.ulcerans_Agy99 VVIADVEEERGESLAAALGADAM---FCRTDVSQPEQVAAVVAAAVDNFG
MAV_0886|M.avium_104 VVIADIETERGERLAAELGGEAV---FRRTDVSDIEQVGALVAAAVEKFG
MAB_4335|M.abscessus_ATCC_1997 VAIGDIDKKLCENTAAEIGNSAI---GLPLDVTDHGSFEAFFDTIAATVG
Mvan_3101|M.vanbaalenii_PYR-1 VWIADRQFGVAEELAQRLRDTGARVHAVELDVRDPAAFATAIAATVQTAG
: : . ** : . . *
Mflv_1267|M.gilvum_PYR-GCK RLDLAFLNAGVTIDWSGDIGSLDVAQYRRSVGVNVDGVVFGVRAAVRTMR
MSMEG_6623|M.smegmatis_MC2_155 GVDVAFLNAGITIAWSGDIGALDLTDYRRSVGVNLDGVVYGVVAAVRTMR
TH_1621|M.thermoresistible__bu GVDLAFLNAGITIEWSGDIAELDLAQYRRSVGVNLDGVVFGARAAVRSMR
Mb2882c|M.bovis_AF2122/97 RIDIAFNNAGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAALRHMV
Rv2857c|M.tuberculosis_H37Rv RIDIAFNNAGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAALRHMV
MLBr_01807|M.leprae_Br4923 PVEVLVANAGITS--DMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMI
MMAR_4765|M.marinum_M GLHVMVNNAGVSGVMHRRFLDDDLADFHRVMAVNVLGVMAGTRDAARHMA
MUL_0336|M.ulcerans_Agy99 GLHVMVNNAGVSGAMHRRFLDDDLADFHRVMAVNVLGVMAGTRDAARHMA
MAV_0886|M.avium_104 GLHVMVNNAGISSPLRR-LLDDDLADFHRVMGVNVLGVMAGTRDAARHMA
MAB_4335|M.abscessus_ATCC_1997 PVDVIVNNAGIMP--ITPFDDESLESIQRQLDINVRGVMWGSQLAIKRMK
Mvan_3101|M.vanbaalenii_PYR-1 RIDYLFNNAGIGIG--GEVDTLTLDDWNDIIDVNLRGVVHGIQAAYPIMI
:. . ***: . . :*: .* * *
Mflv_1267|M.gilvum_PYR-GCK SLGAGTDRAILATASLAGLMPWHPDPVYSLGKHGVVGFMRSVAPNLAAES
MSMEG_6623|M.smegmatis_MC2_155 SR-PGADRAILATASLAGILPWHPDPVYSLGKHGVVGLMRSVAPNLAAEG
TH_1621|M.thermoresistible__bu VRTRRSHGVILATASLAGLIPWHPDPVYSLGKHAVVGFMRSIAPNLAPEG
Mb2882c|M.bovis_AF2122/97 LAGKGSIVNTASFVAVMGSATSQIS--YTASKGGVLAMSRELGVQFARQG
Rv2857c|M.tuberculosis_H37Rv LAGKGSIVNTASFVAVMGSATSQIS--YTASKGGVLAMSRELGVQFARQG
MLBr_01807|M.leprae_Br4923 KQRFGRYIFMGSVVGLSG-VPGQAN--YAASKSGLIGMARSLARELGSRS
MMAR_4765|M.marinum_M AHGGGSIVNLTSIGGIQA-GGGVMT--YRASKAAVIQFTKSAAIELAHYE
MUL_0336|M.ulcerans_Agy99 AHGGGSIVNLTSIGGIQA-GGGVMT--YRASKGAVIQFTKSAAIELAHYE
MAV_0886|M.avium_104 DNGGGAIINLTSIGGIQA-GGGVMT--YRASKAAVIQFTKAAAIELARYD
MAB_4335|M.abscessus_ATCC_1997 PRGGGVIVNIASAAGKAG-FPGLAT--YCATKHAVVGLSESLSLEYEPAG
Mvan_3101|M.vanbaalenii_PYR-1 RQGSGHIVNTASMAGLVT-TSAQAG--YSATKHAVVALSKTMRVEAATHG
: . * * .:: : . :
Mflv_1267|M.gilvum_PYR-GCK IAVHTICPGITETGVLGD-RRPLVERIGVPVMEP-----DAIAEAVMVAL
MSMEG_6623|M.smegmatis_MC2_155 IAVHTICPGITETGVLGD-RRPLVERIGVPVMEP-----EAIADAVMAAV
TH_1621|M.thermoresistible__bu IAVHTICPGITETGVLGD-RRPLVERIGVPVMEA-----DTVSDAVLAAA
Mb2882c|M.bovis_AF2122/97 IRVNALCPGPVNTPLLQE-LFAKNPERAARRM---------VHVPLGRFA
Rv2857c|M.tuberculosis_H37Rv IRVNALCPGPVNTPLLQE-LFAKNPERAARRM---------VHVPLGRFA
MLBr_01807|M.leprae_Br4923 ITANVVAPGFIDTEMTRA-MDSKYQEAALQ------------AIPLGRIG
MMAR_4765|M.marinum_M IRVNAIAPGNIPTPLLAS-SAAGMDQEQVERFTAQIRQTMREDRPLKREG
MUL_0336|M.ulcerans_Agy99 IRVNAIAPGNIPTPLLAS-SAAGLDQEQVERFTAQIRQTMREDRPLKREG
MAV_0886|M.avium_104 IRVNAIAPGNIPTPILGK-SAGDMDPEQRERFEARIREGMREDRPLKREG
MAB_4335|M.abscessus_ATCC_1997 ISVVCVMPGMVNTELISG-LDTHWLLKTVEPEDIANAVVKAVRRGVFPVF
Mvan_3101|M.vanbaalenii_PYR-1 VRVSVLCPGVVRTPILSGGAYGRNKSVSREDLVKLGEALRPMDADDFADR
: . : ** * :
Mflv_1267|M.gilvum_PYR-GCK DAPADATGTCWVAQY---------GKPAWPMVFAPVESPDSRLNVPVRLR
MSMEG_6623|M.smegmatis_MC2_155 AAPLADTGACWVVQH---------GKAPTPMRFADVDCPDARLNVPVRR-
TH_1621|M.thermoresistible__bu RAPIGQTGACWVTQH---------GKPAWPMTFPDVPGPDSKLNVPVRRT
Mb2882c|M.bovis_AF2122/97 EPDEIAAAVAFLASD---------DASFITASTFLVDGGISSAYVTPL--
Rv2857c|M.tuberculosis_H37Rv EPDEIAAAVAFLASD---------DASFITASTFLVDGGISSAYVTPL--
MLBr_01807|M.leprae_Br4923 AVEEIAGAVSFLASE---------DAGYISGAVIPVDGGLGMGH------
MMAR_4765|M.marinum_M TPEDIAEAALYFAGE---------RSRYVTGTVLPVDGGTVAGKVIRRSR
MUL_0336|M.ulcerans_Agy99 TPEDIAEAALYFAGE---------RSRYVTGTVLPVDGGTVAGKVIRRSR
MAV_0886|M.avium_104 TPDDVAEAALYFATE---------RSRYVTGTVLPVDGGTSAGKAMRSKR
MAB_4335|M.abscessus_ATCC_1997 VPKMFGGMYRTMSVLPRSTGKFLTRKLGIQDFMLNAHGSDARKAYEDRAS
Mvan_3101|M.vanbaalenii_PYR-1 ALRAVLRNEAIIVVPRWWKALWYLERLSPALSMRTAAAALKRTREVQSAV
. .
Mflv_1267|M.gilvum_PYR-GCK -------
MSMEG_6623|M.smegmatis_MC2_155 -------
TH_1621|M.thermoresistible__bu SG-----
Mb2882c|M.bovis_AF2122/97 -------
Rv2857c|M.tuberculosis_H37Rv -------
MLBr_01807|M.leprae_Br4923 -------
MMAR_4765|M.marinum_M -------
MUL_0336|M.ulcerans_Agy99 -------
MAV_0886|M.avium_104 QG-----
MAB_4335|M.abscessus_ATCC_1997 HSGPSTE
Mvan_3101|M.vanbaalenii_PYR-1 PPQ----