For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ELQDRVALVTGGGSGIGRAAAQRLAAHGMRVCVVDIDGDAAREVAEPLGGLALTCDVSDPDQVNVTFDRC ATELGGVDVAFLNAGITIAWSGDIGALDLTDYRRSVGVNLDGVVYGVVAAVRTMRSRPGADRAILATASL AGILPWHPDPVYSLGKHGVVGLMRSVAPNLAAEGIAVHTICPGITETGVLGDRRPLVERIGVPVMEPEAI ADAVMAAVAAPLADTGACWVVQHGKAPTPMRFADVDCPDARLNVPVRR*
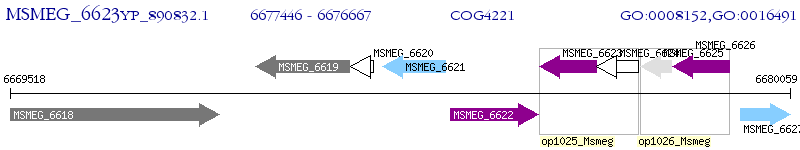
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6623 | - | - | 100% (259) | dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2598 | - | 4e-26 | 34.69% (245) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5235 | - | 9e-25 | 37.70% (191) | short chain dehydrogenase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2882c | - | 2e-26 | 35.87% (223) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1267 | - | 1e-110 | 76.06% (259) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2857c | - | 2e-26 | 35.87% (223) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_02565 | fabG | 7e-18 | 29.84% (248) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. abscessus ATCC 19977 | MAB_4335 | - | 6e-24 | 34.98% (223) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_0924 | - | 1e-23 | 32.43% (259) | carveol-like dehydrogenase |
| M. avium 104 | MAV_3714 | - | 6e-27 | 36.67% (240) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_1621 | - | 1e-106 | 73.08% (260) | dehydrogenase |
| M. ulcerans Agy99 | MUL_0676 | - | 1e-24 | 32.82% (259) | carveol-like dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0852 | - | 2e-25 | 38.05% (226) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3714|M.avium_104 --------------------------------------------------
MSMEG_6623|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1267|M.gilvum_PYR-GCK --------------------------------------------------
TH_1621|M.thermoresistible__bu --------------------------------------------------
MMAR_0924|M.marinum_M --------------------------------------------------
MUL_0676|M.ulcerans_Agy99 --------------------------------------------------
Mvan_0852|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 MAPKCSSDLFSQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLI
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3714|M.avium_104 --------------------------------------------------
MSMEG_6623|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1267|M.gilvum_PYR-GCK --------------------------------------------------
TH_1621|M.thermoresistible__bu --------------------------------------------------
MMAR_0924|M.marinum_M --------------------------------------------------
MUL_0676|M.ulcerans_Agy99 --------------------------------------------------
Mvan_0852|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GGDGRLVEPLRATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELK
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3714|M.avium_104 --------------------------------------------------
MSMEG_6623|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1267|M.gilvum_PYR-GCK --------------------------------------------------
TH_1621|M.thermoresistible__bu --------------------------------------------------
MMAR_0924|M.marinum_M --------------------------------------------------
MUL_0676|M.ulcerans_Agy99 --------------------------------------------------
Mvan_0852|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GLHNFFTPLLRNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGK
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_3714|M.avium_104 --------------------------------------------------
MSMEG_6623|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1267|M.gilvum_PYR-GCK --------------------------------------------------
TH_1621|M.thermoresistible__bu --------------------------------------------------
MMAR_0924|M.marinum_M --------------------------------------------------
MUL_0676|M.ulcerans_Agy99 --------------------------------------------------
Mvan_0852|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_4335|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 ELRRGATVALVYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAAD
Mb2882c|M.bovis_AF2122/97 ---MMDLSQRLAGRVAVITGGGSGIGLAAGRRMRAEGATIVVGDVDV---
Rv2857c|M.tuberculosis_H37Rv ---MMDLSQRLAGRVAVITGGGSGIGLAAGRRMRAEGATIVVGDVDV---
MAV_3714|M.avium_104 ---MMDLTQRLAGRVAVITGAGGGIGLAAARRMHAEGATIVVADIDA---
MSMEG_6623|M.smegmatis_MC2_155 --------MELQDRVALVTGGGSGIGRAAAQRLAAHGMRVCVVDIDG---
Mflv_1267|M.gilvum_PYR-GCK --------MNLAGRVALVTGGASGIGRATAVRLAAEGMQVCVVDIDG---
TH_1621|M.thermoresistible__bu --------VRLDGRAALVTGGGSGIGKATAQRLAAAGMRVCVADIDA---
MMAR_0924|M.marinum_M -------MGALDGRVALITGGARGQGRAHALALAGQGADIALADAPGPMA
MUL_0676|M.ulcerans_Agy99 -------MGALDGRVALITGGARGQGRAHALALAGKGADIALADAPGPMA
Mvan_0852|M.vanbaalenii_PYR-1 -----MVSNHFRGKVCVVTGAASGIGLAVGAALLDHGATVVLADFDADR-
MAB_4335|M.abscessus_ATCC_1997 ----MGTAVQLAGKVVAVTGGARGIGRAIATAFAAEGAKVAIGDIDK---
MLBr_02565|M.leprae_Br4923 STPPTNWDRPLENKVAIVTGAARGIGAAIAEVVARDGARVVAIDVES---
: .:. :**.. * * * . . * : *
Mb2882c|M.bovis_AF2122/97 ------------EAGGAAADELSG--LFVPTDVCDEDAVNGLFDGAAETY
Rv2857c|M.tuberculosis_H37Rv ------------EAGGAAADELSG--LFVPTDVCDEDAVNGLFDGAAETY
MAV_3714|M.avium_104 ------------DAGAAAADELSG--LFVPTDVADEDAVNALFDTAVRRY
MSMEG_6623|M.smegmatis_MC2_155 ------------DAAREVAEPLGG--LALTCDVSDPDQVNVTFDRCATEL
Mflv_1267|M.gilvum_PYR-GCK ------------SAAEEVAESFGG--IGLRCDVTDPDQVDATFAACAAGF
TH_1621|M.thermoresistible__bu ------------DAAAEVADTVNG--VAVRCDVSDPAQVDRVFAACTHEL
MMAR_0924|M.marinum_M ELTYPLGSEEDLLATAELVGQLGRRCLPMVVDVRDAAQVNTAVERTVREL
MUL_0676|M.ulcerans_Agy99 ELTYPLGSEEDLLATAELVGQLGRRCLPMVVDVRDPAQVNTAVERTVREL
Mvan_0852|M.vanbaalenii_PYR-1 -----------LTTVAEGLCMHDGRVHSAVVDVTVQEAVQALVEGAARAH
MAB_4335|M.abscessus_ATCC_1997 ------------KLCENTAAEIGNSAIGLPLDVTDHGSFEAFFDTIAATV
MLBr_02565|M.leprae_Br4923 ----------AAEALDETANRVGG--TTLSLDVTADDAVDKITEHLHDYQ
. ** .:
Mb2882c|M.bovis_AF2122/97 G-RIDIAFNNAGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAALRH
Rv2857c|M.tuberculosis_H37Rv G-RIDIAFNNAGISPPEDNLIENTELAAWQRVQDVNLKSVYLCCRAALRH
MAV_3714|M.avium_104 G-RIDIAFNNAGISPPDDDVIENTEPPAWQRVQDVNLKSVYLCCRAALRH
MSMEG_6623|M.smegmatis_MC2_155 G-GVDVAFLNAGITIAWSGDIGALDLTDYRRSVGVNLDGVVYGVVAAVRT
Mflv_1267|M.gilvum_PYR-GCK G-RLDLAFLNAGVTIDWSGDIGSLDVAQYRRSVGVNVDGVVFGVRAAVRT
TH_1621|M.thermoresistible__bu G-GVDLAFLNAGITIEWSGDIAELDLAQYRRSVGVNLDGVVFGARAAVRS
MMAR_0924|M.marinum_M G-SLDIVLANAGIVS--TGRLEEVSDQVWQQLMDTNLTGVFHTLRAAIPV
MUL_0676|M.ulcerans_Agy99 G-SLDIVLANAGIVS--TGRLEEVSDQVWQQLMDTNLTGVFHTLRAAIPV
Mvan_0852|M.vanbaalenii_PYR-1 G-SLDFLFNNAGVGG--TMPIGVATLEQWRRIVDLNLWGVIYGVHAAVPI
MAB_4335|M.abscessus_ATCC_1997 G-PVDVIVNNAGIMP--ITPFDDESLESIQRQLDINVRGVMWGSQLAIKR
MLBr_02565|M.leprae_Br4923 GGHADILVNNAGITR--DKLLANMDDCRWDAVLEVNLLAPQRLTEGLVGN
* *. . ***: : *: . :
Mb2882c|M.bovis_AF2122/97 MVLA---GKGSIVNTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFA
Rv2857c|M.tuberculosis_H37Rv MVLA---GKGSIVNTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFA
MAV_3714|M.avium_104 MVSA---RRGSIINTASFVAVMGSATSQISYTAAKGGVLALSRELGVQFA
MSMEG_6623|M.smegmatis_MC2_155 MRSR-PGADRAILATASLAGILP-WHPDPVYSLGKHGVVGLMRSVAPNLA
Mflv_1267|M.gilvum_PYR-GCK MRSLGAGTDRAILATASLAGLMP-WHPDPVYSLGKHGVVGFMRSVAPNLA
TH_1621|M.thermoresistible__bu MRVRTRRSHGVILATASLAGLIP-WHPDPVYSLGKHAVVGFMRSIAPNLA
MMAR_0924|M.marinum_M MRQQ---RFGRIVATSSMGGRMG-IPELAAYNATKWGIIGLIKSVALEVA
MUL_0676|M.ulcerans_Agy99 MRQQ---RFGRIVATSSMGGRMG-IPELAAYNATKWGIIGLIKSVALEVA
Mvan_0852|M.vanbaalenii_PYR-1 MVRQ---GSGHIVNTASLAGLVP-FPYQSLYCTTKYGVVGLSESLRYELA
MAB_4335|M.abscessus_ATCC_1997 MKPR---GGGVIVNIASAAGKAG-FPGLATYCATKHAVVGLSESLSLEYE
MLBr_02565|M.leprae_Br4923 GSIS---KGGRVIVLSSMAGIAG-NRGQTNYATAKAGVIGLTQALAPGLA
:: :* . * * .::.: . :
Mb2882c|M.bovis_AF2122/97 RQGIRVNALCPGPVNTPLLQELFAKN--------PERAARRMVHVPLGRF
Rv2857c|M.tuberculosis_H37Rv RQGIRVNALCPGPVNTPLLQELFAKN--------PERAARRMVHVPLGRF
MAV_3714|M.avium_104 RQGIRVNALCPGPVNTPLLQELFAKD--------PQRAARRLVHVPVGRF
MSMEG_6623|M.smegmatis_MC2_155 AEGIAVHTICPGITET-----GVLGD--------RRPLVERIGVPVMEPE
Mflv_1267|M.gilvum_PYR-GCK AESIAVHTICPGITET-----GVLGD--------RRPLVERIGVPVMEPD
TH_1621|M.thermoresistible__bu PEGIAVHTICPGITET-----GVLGD--------RRPLVERIGVPVMEAD
MMAR_0924|M.marinum_M KEGITANVICPTTTQTPMVQPAGIGD--------DQEVPDDLVRRMMKAN
MUL_0676|M.ulcerans_Agy99 KEGITANVICPTTTQTPMMQPAGIGD--------DQEVPDDLVRRMMKAN
Mvan_0852|M.vanbaalenii_PYR-1 ADGVRVSVVCPGNVVSRIFGTPILGRPVDAAPPEDSIPAEEAARLILAGV
MAB_4335|M.abscessus_ATCC_1997 PAGISVVCVMPGMVNTELISGLDTHWLL------KTVEPEDIANAVVKAV
MLBr_02565|M.leprae_Br4923 EKGITINAVAPGFIETQMTAAIPLAT---------REVGRRLNSLLQGGL
.: : * :
Mb2882c|M.bovis_AF2122/97 AEPDEIAAAV--------AFLASDD-------ASFITASTFLVDGGISSA
Rv2857c|M.tuberculosis_H37Rv AEPDEIAAAV--------AFLASDD-------ASFITASTFLVDGGISSA
MAV_3714|M.avium_104 AEPGEIAAAA--------AFLASDD-------ASFITASTFLVDGGISAA
MSMEG_6623|M.smegmatis_MC2_155 AIADAVMAAVAAPLADTGACWVVQH-------GKAPTPMRFADVDCPDAR
Mflv_1267|M.gilvum_PYR-GCK AIAEAVMVALDAPADATGTCWVAQY-------GKPAWPMVFAPVESPDSR
TH_1621|M.thermoresistible__bu TVSDAVLAAARAPIGQTGACWVTQH-------GKPAWPMTFPDVPGPDSK
MMAR_0924|M.marinum_M PIPQPWLQPEDVSR---GVVYLVTD-------PGVITGSVLEVGLGGSAR
MUL_0676|M.ulcerans_Agy99 PIPQPWLQPEDVSR---GVVYLVTD-------PGVITGSVLEVGLGGSAR
Mvan_0852|M.vanbaalenii_PYR-1 AAGEGIIAFPEKPRELWRSYAESPDSV-EGYLKEVARQRRAAFATGDAQA
MAB_4335|M.abscessus_ATCC_1997 RRGVFPVFVPKMFGGMYRTMSVLPRSTGKFLTRKLGIQDFMLNAHGSDAR
MLBr_02565|M.leprae_Br4923 PVDVAETIAY---------FAIPAS--------NAVTGNVIRVCGQAMLG
Mb2882c|M.bovis_AF2122/97 YVTPL----------
Rv2857c|M.tuberculosis_H37Rv YVTPL----------
MAV_3714|M.avium_104 YVTPL----------
MSMEG_6623|M.smegmatis_MC2_155 LNVPVRR--------
Mflv_1267|M.gilvum_PYR-GCK LNVPVRLR-------
TH_1621|M.thermoresistible__bu LNVPVRRTSG-----
MMAR_0924|M.marinum_M MH-------------
MUL_0676|M.ulcerans_Agy99 MH-------------
Mvan_0852|M.vanbaalenii_PYR-1 LFRATQSAGGT----
MAB_4335|M.abscessus_ATCC_1997 KAYEDRASHSGPSTE
MLBr_02565|M.leprae_Br4923 A--------------