For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SLLTGQTAVITGGAQGLGFAIAQRFVDEGARVVLGDVNLEATQEAAEKLGGADVAHAVRCDVTSSAEVDA LVAAAVDTFGALDIMVNNAGITRDATMRKMTEDDFDKVIAVHLKGTWNGLRAAAAIMRENKRGVIVNMSS ISGKVGMIGQTNYSAAKAGIVGMTKAASKELAYLGVRVNAIQPGLIRSAMTEAMPQRIWDSKVAEVPLGR AGEPDEVAKVALFLASDLSSYMTGTVLEVTGGRHL*
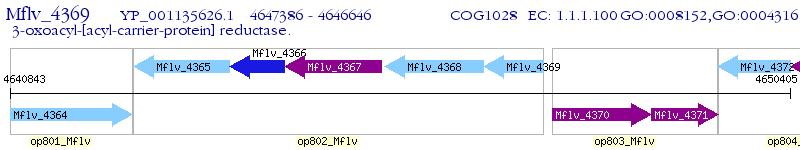
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4369 | fabG | - | 100% (246) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_4505 | - | 3e-44 | 40.00% (240) | short-chain dehydrogenase/reductase SDR |
| M. gilvum PYR-GCK | Mflv_3281 | - | 3e-42 | 40.00% (240) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1385 | fabG | 1e-110 | 80.82% (245) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. tuberculosis H37Rv | Rv1350 | fabG | 1e-110 | 80.82% (245) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 3e-38 | 34.60% (237) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0598 | - | 3e-41 | 40.73% (248) | short chain dehydrogenase |
| M. marinum M | MMAR_4033 | fabG | 1e-111 | 80.89% (246) | 3-oxoacyl- |
| M. avium 104 | MAV_1572 | - | 1e-117 | 85.37% (246) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2206 | fabG | 1e-118 | 86.48% (244) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. thermoresistible (build 8) | TH_3693 | - | 1e-116 | 84.55% (246) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3898 | fabG | 1e-108 | 81.43% (237) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_1975 | fabG | 1e-124 | 91.87% (246) | 3-ketoacyl-(acyl-carrier-protein) reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1385|M.bovis_AF2122/97 --------------MASLLNAR---TAVITGGA-QGLGLAIGQRFVAEGA
Rv1350|M.tuberculosis_H37Rv --------------MASLLNAR---TAVITGGA-QGLGLAIGQRFVAEGA
MMAR_4033|M.marinum_M --------------MSVSLLNG--QIAVVTGGA-QGLGLAIAERFVSEGA
MUL_3898|M.ulcerans_Agy99 --------------MCGAHRDRGYVYPVVTGGA-QGLGLAIAERFVSEGA
MAV_1572|M.avium_104 ---------------MSLLSGQ---TAVITGGA-QGLGFAIAERFVAEGA
Mflv_4369|M.gilvum_PYR-GCK ---------------MSLLTGQ---TAVITGGA-QGLGFAIAQRFVDEGA
Mvan_1975|M.vanbaalenii_PYR-1 ---------MRMGEQVSLLTGQ---TAVITGGA-QGLGFAIAQRFVEEGA
TH_3693|M.thermoresistible__bu ---------------VSLLAGQ---TAVVTGGA-RGLGYAIAERFIAEGA
MSMEG_2206|M.smegmatis_MC2_155 -----------------MLTGQ---TAVVTGGA-QGLGLAIAKRFISEGA
MAB_0598|M.abscessus_ATCC_1997 ---MNLAEAPTEIEGHGLLKGK---AVVVTAAAGTGIGSSTARRALVEGA
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSR---SILVTGGN-RGIGLAVARRLVADGH
::*.. *:* : ..* : :*
Mb1385|M.bovis_AF2122/97 RVVLGDVNLEATEVAAKRLG--GDDVALAVRCDVTQADDVDILIRTAVER
Rv1350|M.tuberculosis_H37Rv RVVLGDVNLEATEVAAKRLG--GDDVALAVRCDVTQADDVDILIRTAVER
MMAR_4033|M.marinum_M RVVLGDVNLEATEAAAKQLG--GGEVAVAVRCDVTKADEVETLLQTALER
MUL_3898|M.ulcerans_Agy99 RVVLGDVNLEATEAAAKQLG--GGEVAVAVRCDVTKADEVETLLQAALER
MAV_1572|M.avium_104 RVVLGDVNLEATQTAAKQLG--GDQVALAVRCDVTKSSEVETLIQTAVER
Mflv_4369|M.gilvum_PYR-GCK RVVLGDVNLEATQEAAEKLG--GADVAHAVRCDVTSSAEVDALVAAAVDT
Mvan_1975|M.vanbaalenii_PYR-1 RVVLGDVNLDATVAAAEKLG--GEDVARAVRCDVTDSAEVDALIAAAVDG
TH_3693|M.thermoresistible__bu RVVIGDIDESATRAAADSLG--GADVAVGMRCDVTRSDEVEALVGAAVKR
MSMEG_2206|M.smegmatis_MC2_155 RVVLGDLNSEATEAAVEELG--GSEVAAAVRCDVTSSADVDALVQAAVER
MAB_0598|M.abscessus_ATCC_1997 DVVVSDYHERRLTETRDELAALGLGKVAAVVCDVTSTEAVDALITESVAA
MLBr_01807|M.leprae_Br4923 RVAVTHR---ASVVPDWFFG---------VECDVTDNDAIGAAFKAVEEH
*.: . . :. : **** : .
Mb1385|M.bovis_AF2122/97 FGGLDVMVNNAGITRDATMRTMTEEQFDQVIAVHLKGTWNGTRLAAAIMR
Rv1350|M.tuberculosis_H37Rv FGGLDVMVNNAGITRDATMRTMTEEQFDQVIAVHLKGTWNGTRLAAAIMR
MMAR_4033|M.marinum_M FGGLDIMVNNAGITRDATMRKMTEEQFDQVIDVHLKGTWNGIRLAAAIMR
MUL_3898|M.ulcerans_Agy99 FGGLDIMVNNAGITRDATMRKMTEEQFDQVIDVHLKGTWNGIRLAAAIMR
MAV_1572|M.avium_104 FGGLDIMVNNAGITRDATMRKMTEEQFDQVIAVHLKGTWNGTRLAAAIMR
Mflv_4369|M.gilvum_PYR-GCK FGALDIMVNNAGITRDATMRKMTEDDFDKVIAVHLKGTWNGLRAAAAIMR
Mvan_1975|M.vanbaalenii_PYR-1 FGSLDIMVNNAGITRDATMRKMTEEQFDQVIAVHLKGTWNGLRAAAAVMR
TH_3693|M.thermoresistible__bu FGSLDVMVNNAGITRDATMRKMTEQQFDEVIAVHLKGTWNGLRAAAAIMR
MSMEG_2206|M.smegmatis_MC2_155 FGGLDIMVNNAGITRDATLRKMTEEQFDQVIAVHLKGTWNGTKAAAAIMR
MAB_0598|M.abscessus_ATCC_1997 LGRIDVLVNNAGLGGETPVADMTDDQWDRVLDVTLTSTFRATRAALRYFR
MLBr_01807|M.leprae_Br4923 QGPVEVLVANAGITSDMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMI
* ::::* ***: : : *::::::.*: * *...:. : * :
Mb1385|M.bovis_AF2122/97 ER-KRGAIVNMSSVSGKVGMVGQTNYSAAKAGIVGMTKAAAKELAHLGIR
Rv1350|M.tuberculosis_H37Rv ER-KRGAIVNMSSVSGKVGMVGQTNYSAAKAGIVGMTKAAAKELAHLGIR
MMAR_4033|M.marinum_M EN-KRGAIVNMSSVSGKVGMVGQTNYSAAKAGIVGMTKAAAKELAHVGVR
MUL_3898|M.ulcerans_Agy99 EN-KRGAIVNMSSVSGKVGMVGQTNYSAAKAGIVGMTKAAAKELAHVGVR
MAV_1572|M.avium_104 EN-KRGAIINMSSVSGKVGMVGQTNYSAAKAGIVGMTKAAAKELAYLGVR
Mflv_4369|M.gilvum_PYR-GCK EN-KRGVIVNMSSISGKVGMIGQTNYSAAKAGIVGMTKAASKELAYLGVR
Mvan_1975|M.vanbaalenii_PYR-1 EQ-KRGAIVNMSSISGKVGMVGQTNYSAAKAGIVGMTKAASKELAYLGVR
TH_3693|M.thermoresistible__bu EQ-KRGAIVNMSSISGKVGMVGQTNYSAAKAGIVGMTKAASKELAYLGVR
MSMEG_2206|M.smegmatis_MC2_155 EN-KRGAIVNMSSISGKVGLIGQTNYSAAKAGIVGMTKAAAKELAYLGVR
MAB_0598|M.abscessus_ATCC_1997 EAGHGGVIVNNASVLGWRAQYGQSHYAAAKAGVMALTRCSAIEAVEHGVR
MLBr_01807|M.leprae_Br4923 KQ-RFGRYIFMGSVVGLSGVPGQANYAASKSGLIGMARSLARELGSRSIT
: : * : .*: * . **::*:*:*:*::.:::. : * .:
Mb1385|M.bovis_AF2122/97 VNAIAPGLIRSAMTEAMP-QRIWDQKLAEVPMGRAGEPSEVASVAVFLAS
Rv1350|M.tuberculosis_H37Rv VNAIAPGLIRSAMTEAMP-QRIWDQKLAEVPMGRAGEPSEVASVAVFLAS
MMAR_4033|M.marinum_M VNAIAPGLIRSAMTEAMP-QRIWDQKLAEVPMGRAGEPSEVASVALFLAC
MUL_3898|M.ulcerans_Agy99 VNAIAPGLIRSAMTEAMP-QRIWDQKLAEVPMGRAGEPSEVASVALFLAC
MAV_1572|M.avium_104 VNAIAPGLIRSAMTEAMP-QRIWDSKVAEVPMGRAGEPSEVASVALFLAS
Mflv_4369|M.gilvum_PYR-GCK VNAIQPGLIRSAMTEAMP-QRIWDSKVAEVPLGRAGEPDEVAKVALFLAS
Mvan_1975|M.vanbaalenii_PYR-1 VNAIQPGLIRSAMTEAMP-QRIWDSKVAEVPMGRAGEPEEVANVALFLAS
TH_3693|M.thermoresistible__bu VNAIQPGLIRSAMTEAMP-QRIWDEKLAEIPMGRAGEPEEVAKVALFLAS
MSMEG_2206|M.smegmatis_MC2_155 VNAIQPGLIRSAMTEAMP-QRIWDEKLAEIPMGRAGEPDEVAKVALFLAS
MAB_0598|M.abscessus_ATCC_1997 VNAVAPSIARHKFLDKVTSSDLLDKLSSDEAFGRAAEPWEVAATIAFLAS
MLBr_01807|M.leprae_Br4923 ANVVAPGFIDTEMTRAMD-SKYQEAALQAIPLGRIGAVEEIAGAVSFLAS
.*.: *.: : : . : .:** . *:* . ***.
Mb1385|M.bovis_AF2122/97 DLSSYMTGTVLDVTGGRFI--
Rv1350|M.tuberculosis_H37Rv DLSSYMTGTVLDVTGGRFI--
MMAR_4033|M.marinum_M DLSSYMTGTVLDVTGGRFI--
MUL_3898|M.ulcerans_Agy99 DLSSYMTGTVLDVTGGRFI--
MAV_1572|M.avium_104 DMSSYMTGTVMEITGGRHL--
Mflv_4369|M.gilvum_PYR-GCK DLSSYMTGTVLEVTGGRHL--
Mvan_1975|M.vanbaalenii_PYR-1 DLSSYMTGTVLEVTGGRHL--
TH_3693|M.thermoresistible__bu DLSSYMTGTVLEVTGGRHL--
MSMEG_2206|M.smegmatis_MC2_155 DLSSYMTGTVLEVTGGRHV--
MAB_0598|M.abscessus_ATCC_1997 DYSSYLTGEVISVSSQRA---
MLBr_01807|M.leprae_Br4923 EDAGYISGAVIPVDGGLGMGH
: :.*::* *: : .