For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LRGDGPGPARQWSADAVVVGVLAFALGVAAAGRPSLWVDEAGTLSAATRPVDALWTLVHHVDAVHGFYYL LLRGWFEIVPVTEFWARVPSALMVGVAAAGTCVLGSRLSGRKVGIAAGVVFAVLPRTTWAAVEARPYALS MACAVWLTVLFVAAVRRGGRRWWLGYGAALVGAVTVNVMVVLILAGHAAMLAATRPDRRTVRAWLVTVVV ASVALIPLFVVLLSQQAQIAWIWPVGPVTLGQIAGEQYFPSVYSDSVRAVGPDQQQFSAEQLRVAMSAWA RVAPLIVVIVIVAALAVRARRRHGALIENPSGTRVLVLGAVAWIVVPTVVLLGYSVLVRPLYQPHYLTFT TPALALLIGLCVVTLCRDDTRRLTATLAVLAVAAAPNYLAQRGPYAKYGSDYSQVAELLARESGPRDCVV VDDRTAPSVPHAIEGARRAHPVAARDLGLAPPERTALFSARDPLGVRADDLRTCAVLWTIGDAAPGSPAD APVDRVARAQGFVPAERWRFNQTEVVRSVPAGARS*
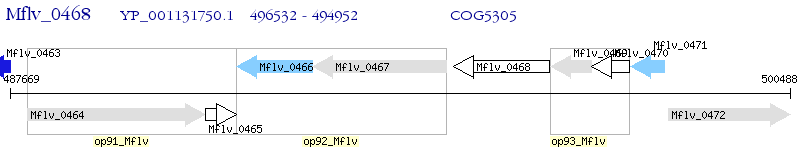
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0468 | - | - | 100% (526) | hypothetical protein Mflv_0468 |
| M. gilvum PYR-GCK | Mflv_2269 | - | 7e-08 | 26.10% (387) | glycosyl transferase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1661c | - | 6e-87 | 38.94% (529) | hypothetical protein Mb1661c |
| M. tuberculosis H37Rv | Rv1635c | - | 6e-87 | 38.94% (529) | transmembrane protein |
| M. leprae Br4923 | MLBr_01389 | - | 2e-89 | 36.83% (543) | hypothetical protein MLBr_01389 |
| M. abscessus ATCC 19977 | MAB_2025 | - | 5e-05 | 24.72% (271) | purine/cytosine permease |
| M. marinum M | MMAR_2439 | - | 6e-92 | 37.88% (528) | transmembrane protein |
| M. avium 104 | MAV_4843 | - | 2e-96 | 40.57% (530) | hypothetical protein MAV_4843 |
| M. smegmatis MC2 155 | MSMEG_4988 | - | 9e-08 | 25.24% (317) | integral membrane protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1618 | - | 2e-92 | 38.07% (528) | transmembrane protein |
| M. vanbaalenii PYR-1 | Mvan_0891 | - | 1e-06 | 28.57% (245) | putative integral membrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1661c|M.bovis_AF2122/97 -----------MHASRPGAPPHAG----LPSRRTAGDQDHRADPKVTRIM
Rv1635c|M.tuberculosis_H37Rv -----------MHASRPGAPPHAG----LPSRRTAGDQDHRADPKVTRIM
MMAR_2439|M.marinum_M MEFQAAQHFSRLWAAQPGGPPKPRGQRPPNPRKMTRDQDHRWALTVTSIM
MUL_1618|M.ulcerans_Agy99 -------------------------------------------------M
MLBr_01389|M.leprae_Br4923 ---------------------------------------------MTSIM
MAV_4843|M.avium_104 -------------------------------------------------M
Mflv_0468|M.gilvum_PYR-GCK -------------------------------------------------M
MSMEG_4988|M.smegmatis_MC2_155 ------------------------------------------MTLVARDP
Mvan_0891|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2025|M.abscessus_ATCC_1997 --------------------------------------------------
Mb1661c|M.bovis_AF2122/97 SASTLEQPAAAH-VDELVARMRGRLLDPLAIA--VLAAVISGAWASRPSL
Rv1635c|M.tuberculosis_H37Rv SASTLEQPAAAH-VDELVARMRGRLLDPLAIA--VLAAVISGAWASRPSL
MMAR_2439|M.marinum_M STPTLEQGATAT-DKAERVRPRGRRLDPWLMA--LLATLISVAWAGKPSL
MUL_1618|M.ulcerans_Agy99 STPTLEQGATAT-DKAERVRPRGRRLDPWLMA--LLATLISVAWAGRPSL
MLBr_01389|M.leprae_Br4923 SVSALEQSAADVGDNSARQHAHGALPDSLAIA--MLATVISGAWASRPSL
MAV_4843|M.avium_104 SSPTRPAVPAAL--------------DPWIVA--ALAAAVSLAGAARPSF
Mflv_0468|M.gilvum_PYR-GCK LRGDGPGPARQW------------SADAVVVG--VLAFALGVAAAGRPSL
MSMEG_4988|M.smegmatis_MC2_155 ARAEVEAPDSRTSESVRPRSRAERVALPLLLAGTALLYLWNLSISGWANP
Mvan_0891|M.vanbaalenii_PYR-1 -----------------------------------------MPIGRKEAV
MAB_2025|M.abscessus_ATCC_1997 -------------------------MDEIQACCSATSLIASRESVGRDRV
Mb1661c|M.bovis_AF2122/97 WFDEGATISASASRTLPELWSLLGHIDAVHG--LYYLLMHGWFAIFPPTE
Rv1635c|M.tuberculosis_H37Rv WFDEGATISASASRTLPELWSLLGHIDAVHG--LYYLLMHGWFAIFPPTE
MMAR_2439|M.marinum_M WFDEGATISAAANRTLPELWKLLGHIDAVHG--FYYLLMHGWYAIFPPTE
MUL_1618|M.ulcerans_Agy99 WFDEGATISAAANRTLPELWKLLGHIDAVHG--FYYLLMHGWYAIFPPTE
MLBr_01389|M.leprae_Br4923 WFDEAATISASASRTVPELWRLLSHIDAVHG--LYYLLMHGWFAIFPSTE
MAV_4843|M.avium_104 WYDEAATISASYSRSLTQMWHMLGNVDAVHG--LYYLLMHGWFRLVPPTE
Mflv_0468|M.gilvum_PYR-GCK WVDEAGTLSAAT-RPVDALWTLVHHVDAVHG--FYYLLLRGWFEIVPVTE
MSMEG_4988|M.smegmatis_MC2_155 FYSAAAQAGASDWIAMLFGSSDPANAITVDKTPAALWVIDLSVRLFGLSS
Mvan_0891|M.vanbaalenii_PYR-1 AVAVGSVLVVAAFVIPHLDLGVVTPLINATP-----ERLRSFADTAPIFG
MAB_2025|M.abscessus_ATCC_1997 GAIERAGIDYLPEAARGSSPKNLFAVFVGGN-LAFSTIIFGWLPIVMGLT
. :
Mb1661c|M.bovis_AF2122/97 LWSRLPS---CLAIGAAAAGVVVFAKQFSGRTTAVCAGAVFAILPRVTWA
Rv1635c|M.tuberculosis_H37Rv LWSRLPS---CLAIGAAAAGVVVFAKQFSGRTTAVCAGAVFAILPRVTWA
MMAR_2439|M.marinum_M FWSRVPS---SLAIGGAAAGVVVFTKQFSGRSTALCAGAVFAILPRVTWA
MUL_1618|M.ulcerans_Agy99 FWSRVPS---SLAIGGAAAGVVVFTKQFSGRSTALCAGAVFAILPRVTWA
MLBr_01389|M.leprae_Br4923 FWSRVPS---CLAIGAAAAGVTVFTRQFATRTTAVYAGIVFAILPRITWA
MAV_4843|M.avium_104 FWSRAPS---GLAVGAAAAGVVVLGRQFSSRTVAVASGVFCAVLPRTTWA
Mflv_0468|M.gilvum_PYR-GCK FWARVPS---ALMVGVAAAGTCVLGSRLSGRKVGIAAGVVFAVLPRTTWA
MSMEG_4988|M.smegmatis_MC2_155 WSVLLPQ---ALEGIAAVAVLYAAVRRAAGWHAAVLAGAVLALTPVAALI
Mvan_0891|M.vanbaalenii_PYR-1 WWN--------THVGAGTIPAFAIGAAAVLWGPPLAQRLPWRWLPWLSWT
MAB_2025|M.abscessus_ATCC_1997 FWQAVSSSVTGLLIGLMLTAPMSLFGPRTGTNNPVSSGAHFGVRGRLIGS
:
Mb1661c|M.bovis_AF2122/97 GIEARSSALSVAAAVWLTVLLVAAVRCNTQRRWLLYALVLMLS--ILVSI
Rv1635c|M.tuberculosis_H37Rv GIEARSSALSVAAAVWLTVLLVAAVRCNTQRRWLLYALVLMLS--ILVSI
MMAR_2439|M.marinum_M GIEARSSALSVAAAVWLTVLLVAAVRRNRPWLWLLYALVLMVS--ILINI
MUL_1618|M.ulcerans_Agy99 GIEARSSALSVAAAVWLTVLLVAAVRRDRPWLWLLYALVLMVS--ILINI
MLBr_01389|M.leprae_Br4923 GIEARSSALSVAAAMWLTVLLVASVQRNRPRLWLCYALTLMLS--ILLNL
MAV_4843|M.avium_104 GVEARPYALSMMAAVWLSALLVFAARRQSRWPWLCFGLALVCS--VLLDA
Mflv_0468|M.gilvum_PYR-GCK AVEARPYALSMACAVWLTVLFVAAVRRGGRRWWLGYGAALVGA--VTVNV
MSMEG_4988|M.smegmatis_MC2_155 FRFNNPDALLVLLLVVAAYCVQRGIEKDASRWWFVAAGAAVGFGFLAKML
Mvan_0891|M.vanbaalenii_PYR-1 VSCAWAFSLAMVDGWQR----GFAGRLTARHEYLRQVPTVTDIPEALRTF
MAB_2025|M.abscessus_ATCC_1997 SLTLMFALVYAAIAVWTSGEALIAGASRLFGAPQNHVAFLLGYGVIALEV
: .
Mb1661c|M.bovis_AF2122/97 NLALLVPAYATMVPLLASGKSRKSPVIWWTVVTAAALGAMTPFILFAHGQ
Rv1635c|M.tuberculosis_H37Rv NLALLVPAYATMVPLLASGKSRKSPVIWWTVVTAAALGAMTPFILFAHGQ
MMAR_2439|M.marinum_M NLGLLVPVYGALLPLLAPKKSRKSPGIWWAVTSVVAIGAMTPFVLFAHGQ
MUL_1618|M.ulcerans_Agy99 NLGLLVPVYGALLPLLAPKKSRKSPGIWWAVTSVVAIGAMTPFVLFAHGQ
MLBr_01389|M.leprae_Br4923 TLATLVLVYAVILPWLAPNKFRNSPFIWWAVTSVVALGTITPFILFAHGQ
MAV_4843|M.avium_104 YIALLLAAYAVFVGVCCRTR---TVLWRFGISSVVAVGVLLPFLLTVAGQ
Mflv_0468|M.gilvum_PYR-GCK MVVLILAGHAAMLAATRPDR---RTVRAWLVTVVVASVALIPLFVVLLSQ
MSMEG_4988|M.smegmatis_MC2_155 QAFLVLPAFCAAALLAGDRPLRARIADLAAAGTAMVVSGGWYLVLVELWP
Mvan_0891|M.vanbaalenii_PYR-1 AGRILDYQPDSWITHVSGHPPGALLTFVWLDRIGLGGGAWAGLLCLLAGS
MAB_2025|M.abscessus_ATCC_1997 ILIALYGHATIVWAQKIIIPLAVVVLALGVLAYAPNFHPMGSAQHGYALQ
:
Mb1661c|M.bovis_AF2122/97 VWQVGWIAGLNRNIILDVIH-RQYFD----HSV-----------------
Rv1635c|M.tuberculosis_H37Rv VWQVGWIAGLNRNIILDVIH-RQYFD----HSV-----------------
MMAR_2439|M.marinum_M VWQVGWISGLNRNLFLDVVH-RQYFD----HSV-----------------
MUL_1618|M.ulcerans_Agy99 VWQVGWISGLNRNLFLDVVH-RQYFD----HSV-----------------
MLBr_01389|M.leprae_Br4923 VWQVDWIFRVSWHYVFDITQ-RQYFD----HSV-----------------
MAV_4843|M.avium_104 AHQISWVAPIGHRTIEDVVM-QQYFE----RSP-----------------
Mflv_0468|M.gilvum_PYR-GCK QAQIAWIWPVGPVTLGQIAG-EQYFPSVYSDSVRAVGPDQQQFSAEQLR-
MSMEG_4988|M.smegmatis_MC2_155 AASRPYIGGSQNNSIVELALGYNGFGRLTGAEVGGLGNSNFDVGAGRLFG
Mvan_0891|M.vanbaalenii_PYR-1 SIAAAIVVTVRALADEDTARAAAPFV------------------------
MAB_2025|M.abscessus_ATCC_1997 EFWPTWLFAVTVSVGGPLSYAPSVGDYTRRISR-----------------
:
Mb1661c|M.bovis_AF2122/97 -----PFAILAGLIVAAGIAAHLAGARGPGG--------DTHRLVLVSAA
Rv1635c|M.tuberculosis_H37Rv -----PFAILAGLIVAAGIAAHLAGARGPGG--------DTHRLVLVSAA
MMAR_2439|M.marinum_M -----AFAILAGVLVVAAIAVRLTGIARPGE--------GTRPLLLASIA
MUL_1618|M.ulcerans_Agy99 -----AFAILAGVLVVAAIAVRLTGIARPGE--------GTRPLLLASIA
MLBr_01389|M.leprae_Br4923 -----SFAIATAVIIVPAIATRLAGLRAPAG--------DLRSLVIICTA
MAV_4843|M.avium_104 -----PFAVLSALLICAAIALWLSRSAPLGP--------SERQLLVLATC
Mflv_0468|M.gilvum_PYR-GCK -VAMSAWARVAPLIVVIVIVAALAVRARRRHGALIENPSGTRVLVLGAVA
MSMEG_4988|M.smegmatis_MC2_155 WQMGADIAWLLPAALVCLVAGLIVTRRAPRT-------DRIRAALIIWGG
Mvan_0891|M.vanbaalenii_PYR-1 ------AVAPTAIWIAVSADGYFAGVAAWG--------ITLLALAVRGSV
MAB_2025|M.abscessus_ATCC_1997 ------SRYSDRQIAVAVSAGIFLGLFSTAT-----------FGAFTAST
: .
Mb1661c|M.bovis_AF2122/97 WIVVPTAVVLIYSATVEPIYYPRYLILTAPAAAVILAVCVVTIAR-----
Rv1635c|M.tuberculosis_H37Rv WIVVPTAVVLIYSATVEPIYYPRYLILTAPAAAVILAVCVVTIAR-----
MMAR_2439|M.marinum_M WVVIPTGIVLLYSAIVEPIYYPRYLILTAPAAAVILAVCIVTLAR-----
MUL_1618|M.ulcerans_Agy99 WVVIPTGIVLLYSAIVEPIYYPRYLILTAPAAAVILAVCIVTLAR-----
MLBr_01389|M.leprae_Br4923 WIVIPTTLMVGYSAVIEPVYYPRYLILTAPAAAIVIAVCIVTVAR-----
MAV_4843|M.avium_104 WVGIPTAAIVAYSALVHPIYTPRYLCFTAPAMALILGVCSAAIAA-----
Mflv_0468|M.gilvum_PYR-GCK WIVVPTVVLLGYSVLVRPLYQPHYLTFTTPALALLIGLCVVTLCRD----
MSMEG_4988|M.smegmatis_MC2_155 WLVVTAVVFSYMNGIVHPYYT----VALAPAIGAVIGIGATSMWRHRNDI
Mvan_0891|M.vanbaalenii_PYR-1 RQPVAAAIGSGLLLGWAVFLNYGLVLMAFPAVAVLLGAATWQAAVR----
MAB_2025|M.abscessus_ATCC_1997 FSAPTASYVADMVATAPGWYVVPLLILAVLGGCGQGVISIYSSGLD----
.: .
Mb1661c|M.bovis_AF2122/97 -------KPWLIAGVVFLLAAAAFPNYFFTQRGPYAKEGWDYSQVADVIS
Rv1635c|M.tuberculosis_H37Rv -------KPWLIAGVVFLLAAAAFPNYFFTQRGPYAKEGWDYSQVADVIS
MMAR_2439|M.marinum_M -------KPWLITGVVVLLAVAAFPNYFFTQRGPYAKEGWDYSQVADVIS
MUL_1618|M.ulcerans_Agy99 -------KPWLITGVVVLLAVAAFPNYFFTQRGPYAKEGWDYSQVADVIS
MLBr_01389|M.leprae_Br4923 -------KPWPIAGVLVLFAVAAFPNYLFTQRGRYAKEGWDYSQVADVIS
MAV_4843|M.avium_104 -------KPWVTTAVVGVFAIAAAPNYIRAQRNPYAKYGMDYSQVADLIT
Mflv_0468|M.gilvum_PYR-GCK -------DTRRLTATLAVLAVAAAPNYL-AQRGPYAKYGSDYSQVAELLA
MSMEG_4988|M.smegmatis_MC2_155 RVATAMAAAVAVTVILAAVLLARHNDPMPWLRATVAVVGVAVAVLLLVVD
Mvan_0891|M.vanbaalenii_PYR-1 --------TLVPAVVAALAVVAVFAAAGFWWFDGYLLVQERYWQG---IA
MAB_2025|M.abscessus_ATCC_1997 ------LESIVPRLNRTQTTLIASGIAVVLMYLGVVVTDAIDSVTGMTVV
. :
Mb1661c|M.bovis_AF2122/97 AHAKPGDCLLVDN-TAGWRPGP---IRALLATRPAAFRSLIDVERGTYGP
Rv1635c|M.tuberculosis_H37Rv AHAKPGDCLLVDN-TAGWRPGP---IRALLATRPAAFRSLIDVERGTYGP
MMAR_2439|M.marinum_M AHAAPGDCLLVDN-TVPWKPGP---VRALLATRPAAFRSLIDVERGAYGP
MUL_1618|M.ulcerans_Agy99 THAAPGDCLLVDN-TVPWKPGP---VRALLATRPAAFRSLIDVERGAYGP
MLBr_01389|M.leprae_Br4923 SQAAPGDCLIVDN-TVPWRPGP---IRALLAARPAAFRSLIDIERGFYGP
MAV_4843|M.avium_104 AKAAPGDCLLVND-TVTFMPAP---MRPLLAARPDAYRKLIDLTLWQRAV
Mflv_0468|M.gilvum_PYR-GCK RESGPRDCVVVDDRTAPSVPHA---IEGARRAHPVAAR-----DLGLAPP
MSMEG_4988|M.smegmatis_MC2_155 LVHTRLVRPLAALTVLICLAGPGAYAVATAAAPHTGAIPSVGPSRGFGGG
Mvan_0891|M.vanbaalenii_PYR-1 NDRPFQYWGWANFASVVCAIGLG----SVAGLGRVFDIAAIKRRSGLHLL
MAB_2025|M.abscessus_ATCC_1997 LNSFAGSWVAINICGFFKANRGQYDPKALQRFGQDGRGGLYWFSHGFNVR
Mb1661c|M.bovis_AF2122/97 KVGTLWDGHVAVWLTTAKIDKCPTLWTIANRDKSL--PDHQVGEMLSPGT
Rv1635c|M.tuberculosis_H37Rv KVGTLWDGHVAVWLTTAKIDKCPTLWTIANRDKSL--PDHQVGEMLSPGT
MMAR_2439|M.marinum_M KVGTLWDGHVAVWLTTEKINKCSTLWTISNRDNAL--PDHEVGPALSPGT
MUL_1618|M.ulcerans_Agy99 KVGTLWDGHVAVWLTTEKINKCSTLWTISNRDNAL--PDHEVGPALSPGT
MLBr_01389|M.leprae_Br4923 TVGTLWDGHVPVWLVTAKINKCSTVWTVSDRDTSL--PDHQAGQLLSPGL
MAV_4843|M.avium_104 DRNDVFDTNLIPEVVAGPLSHCAVLWIITQADPSE--PAHQQGPALPPGP
Mflv_0468|M.gilvum_PYR-GCK ERTALFSARDPLGVRADDLRTCAVLWTIGDAAPGS--PAD----------
MSMEG_4988|M.smegmatis_MC2_155 PFSAPTPGEALTATLAADADNYTWAAAVVGSSNAAGYQLATGAPVMAVGG
Mvan_0891|M.vanbaalenii_PYR-1 VLGALLAIVVADLSRLSKAETERIWLPFTVWLVAS---------------
MAB_2025|M.abscessus_ATCC_1997 AVVPFIAGSGLGLLTVQNDLYQAPFANIAGGIDVS---------------
.
Mb1661c|M.bovis_AF2122/97 GFGRTPVYRFPSYLGFRIVERWQFHYSQVVKSTR----------------
Rv1635c|M.tuberculosis_H37Rv GFGRTPVYRFPSYLGFRIVERWQFHYSQVVKSTR----------------
MMAR_2439|M.marinum_M AFGKTPVYLFPHYLGFHIVERWQFHYSQVIKSTR----------------
MUL_1618|M.ulcerans_Agy99 AFGKTPVYLFPHYLGFHIVERWQFHYSQVIKSTR----------------
MLBr_01389|M.leprae_Br4923 ILGRAPAYQFPSYLGFRIVERWQFHYSQVIKSTR----------------
MAV_4843|M.avium_104 VYGATPAFAVPHDLGFRLVERWQFNLVQVFEATR----------------
Mflv_0468|M.gilvum_PYR-GCK ----APVDRVARAQGFVPAERWRFNQTEVVRSVPAGARS-----------
MSMEG_4988|M.smegmatis_MC2_155 FNGTDPAPTLAEFEELVAGRQIHYFIRSRIMGARPGHTPTGSQEAAEIAR
Mvan_0891|M.vanbaalenii_PYR-1 -TALLPPRSARWWLALNVIGALAINHLILTNW------------------
MAB_2025|M.abscessus_ATCC_1997 -----LLLSVVVGGGLYWLAQVSWPAPRVVSSGSSA--------------
:
Mb1661c|M.bovis_AF2122/97 -----------------------------
Rv1635c|M.tuberculosis_H37Rv -----------------------------
MMAR_2439|M.marinum_M -----------------------------
MUL_1618|M.ulcerans_Agy99 -----------------------------
MLBr_01389|M.leprae_Br4923 -----------------------------
MAV_4843|M.avium_104 -----------------------------
Mflv_0468|M.gilvum_PYR-GCK -----------------------------
MSMEG_4988|M.smegmatis_MC2_155 WVQAHFTPITIDDVTIYDLTQRVPNTPIS
Mvan_0891|M.vanbaalenii_PYR-1 -----------------------------
MAB_2025|M.abscessus_ATCC_1997 -----------------------------