For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
EFQAAQHFSRLWAAQPGGPPKPRGQRPPNPRKMTRDQDHRWALTVTSIMSTPTLEQGATATDKAERVRPR GRRLDPWLMALLATLISVAWAGKPSLWFDEGATISAAANRTLPELWKLLGHIDAVHGFYYLLMHGWYAIF PPTEFWSRVPSSLAIGGAAAGVVVFTKQFSGRSTALCAGAVFAILPRVTWAGIEARSSALSVAAAVWLTV LLVAAVRRNRPWLWLLYALVLMVSILININLGLLVPVYGALLPLLAPKKSRKSPGIWWAVTSVVAIGAMT PFVLFAHGQVWQVGWISGLNRNLFLDVVHRQYFDHSVAFAILAGVLVVAAIAVRLTGIARPGEGTRPLLL ASIAWVVIPTGIVLLYSAIVEPIYYPRYLILTAPAAAVILAVCIVTLARKPWLITGVVVLLAVAAFPNYF FTQRGPYAKEGWDYSQVADVISAHAAPGDCLLVDNTVPWKPGPVRALLATRPAAFRSLIDVERGAYGPKV GTLWDGHVAVWLTTEKINKCSTLWTISNRDNALPDHEVGPALSPGTAFGKTPVYLFPHYLGFHIVERWQF HYSQVIKSTR*
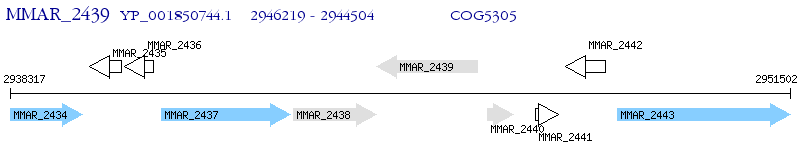
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2439 | - | - | 100% (571) | transmembrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1661c | - | 0.0 | 75.99% (558) | hypothetical protein Mb1661c |
| M. gilvum PYR-GCK | Mflv_0468 | - | 5e-92 | 37.88% (528) | hypothetical protein Mflv_0468 |
| M. tuberculosis H37Rv | Rv1635c | - | 0.0 | 75.99% (558) | transmembrane protein |
| M. leprae Br4923 | MLBr_01389 | - | 0.0 | 70.40% (527) | hypothetical protein MLBr_01389 |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_3138 | - | 0.0 | 71.43% (497) | hypothetical protein MAV_3138 |
| M. smegmatis MC2 155 | MSMEG_0999 | - | 2e-06 | 26.67% (255) | integral membrane protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1618 | - | 0.0 | 99.43% (522) | transmembrane protein |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2439|M.marinum_M MEFQAAQHFSRLWAAQPGGPPKPRGQRPPNPRKMTRDQDHRWALTVTSIM
MUL_1618|M.ulcerans_Agy99 -------------------------------------------------M
Mb1661c|M.bovis_AF2122/97 -----------MHASRPGAPPHAG----LPSRRTAGDQDHRADPKVTRIM
Rv1635c|M.tuberculosis_H37Rv -----------MHASRPGAPPHAG----LPSRRTAGDQDHRADPKVTRIM
MLBr_01389|M.leprae_Br4923 ---------------------------------------------MTSIM
MAV_3138|M.avium_104 --------------------------------------------------
Mflv_0468|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_0999|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_2439|M.marinum_M STPTLEQGATAT-DKAERVRPRGRRLDPWLMALLATLISVAWAGKPSLWF
MUL_1618|M.ulcerans_Agy99 STPTLEQGATAT-DKAERVRPRGRRLDPWLMALLATLISVAWAGRPSLWF
Mb1661c|M.bovis_AF2122/97 SASTLEQPAAAH-VDELVARMRGRLLDPLAIAVLAAVISGAWASRPSLWF
Rv1635c|M.tuberculosis_H37Rv SASTLEQPAAAH-VDELVARMRGRLLDPLAIAVLAAVISGAWASRPSLWF
MLBr_01389|M.leprae_Br4923 SVSALEQSAADVGDNSARQHAHGALPDSLAIAMLATVISGAWASRPSLWF
MAV_3138|M.avium_104 ------------------------MLDPWAIAVLATALSAAWACRPSLWF
Mflv_0468|M.gilvum_PYR-GCK ---MLRGDGPGP--------ARQWSADAVVVGVLAFALGVAAAGRPSLWV
MSMEG_0999|M.smegmatis_MC2_155 -------------------------MQIGRRELVAVIIAILLVTAAFVVP
: ::* :. . . :
MMAR_2439|M.marinum_M DEGATISAAANRTLPELWKLLGHIDAVHGFYYLLMHGWYAIFPPTEFWSR
MUL_1618|M.ulcerans_Agy99 DEGATISAAANRTLPELWKLLGHIDAVHGFYYLLMHGWYAIFPPTEFWSR
Mb1661c|M.bovis_AF2122/97 DEGATISASASRTLPELWSLLGHIDAVHGLYYLLMHGWFAIFPPTELWSR
Rv1635c|M.tuberculosis_H37Rv DEGATISASASRTLPELWSLLGHIDAVHGLYYLLMHGWFAIFPPTELWSR
MLBr_01389|M.leprae_Br4923 DEAATISASASRTVPELWRLLSHIDAVHGLYYLLMHGWFAIFPSTEFWSR
MAV_3138|M.avium_104 DEGATISAAASRTLPELWRLLGHIDAVHGAYYLLMHGWFALFPPTEFFSR
Mflv_0468|M.gilvum_PYR-GCK DEAGTLSAAT-RPVDALWTLVHHVDAVHGFYYLLLRGWFEIVPVTEFWAR
MSMEG_0999|M.smegmatis_MC2_155 HLDLGIVTPLINSTPAQIYSFADTAPIFGWWNAHVG-----------WGT
. : :. .. . . .:.* : : :.
MMAR_2439|M.marinum_M VPSSLAIGGAAAGVVVFTKQFSGRSTALCAGAVFAILPRVTWAGIEARSS
MUL_1618|M.ulcerans_Agy99 VPSSLAIGGAAAGVVVFTKQFSGRSTALCAGAVFAILPRVTWAGIEARSS
Mb1661c|M.bovis_AF2122/97 LPSCLAIGAAAAGVVVFAKQFSGRTTAVCAGAVFAILPRVTWAGIEARSS
Rv1635c|M.tuberculosis_H37Rv LPSCLAIGAAAAGVVVFAKQFSGRTTAVCAGAVFAILPRVTWAGIEARSS
MLBr_01389|M.leprae_Br4923 VPSCLAIGAAAAGVTVFTRQFATRTTAVYAGIVFAILPRITWAGIEARSS
MAV_3138|M.avium_104 LPSALAVGAAAAGVTVFTRQFAPRRTAVCAGAVFALLPRMTWAGMEARPY
Mflv_0468|M.gilvum_PYR-GCK VPSALMVGVAAAGTCVLGSRLSGRKVGIAAGVVFAVLPRTTWAAVEARPY
MSMEG_0999|M.smegmatis_MC2_155 VPAVLIG----AATVWWGPAVAQR-------LPWRALPLVAWVTSALWAF
:*: * *.. .: * : ** :*. .
MMAR_2439|M.marinum_M ALSVAAAVWLTVLLVAAVRRNRPWLWLLYALVLMVSILININLGLLVPVY
MUL_1618|M.ulcerans_Agy99 ALSVAAAVWLTVLLVAAVRRDRPWLWLLYALVLMVSILININLGLLVPVY
Mb1661c|M.bovis_AF2122/97 ALSVAAAVWLTVLLVAAVRCNTQRRWLLYALVLMLSILVSINLALLVPAY
Rv1635c|M.tuberculosis_H37Rv ALSVAAAVWLTVLLVAAVRCNTQRRWLLYALVLMLSILVSINLALLVPAY
MLBr_01389|M.leprae_Br4923 ALSVAAAMWLTVLLVASVQRNRPRLWLCYALTLMLSILLNLTLATLVLVY
MAV_3138|M.avium_104 AFVAAAAVWLTVLFVAAVRRGAPRGWVGYALALMLAILLNLNMVLMVPVY
Mflv_0468|M.gilvum_PYR-GCK ALSMACAVWLTVLFVAAVRRGGRRWWLGYGAALVGAVTVNVMVVLILAGH
MSMEG_0999|M.smegmatis_MC2_155 SLAMIDG--WQRGFAGRLTARHEYLRQVPSVTDIGEALRTFSSRILDFQP
:: . :.. : . . : .. :
MMAR_2439|M.marinum_M GALLPLLAPKKSRKSPGIWWAVTSVVAIGAMTPFVLFAHGQVWQVGWISG
MUL_1618|M.ulcerans_Agy99 GALLPLLAPKKSRKSPGIWWAVTSVVAIGAMTPFVLFAHGQVWQVGWISG
Mb1661c|M.bovis_AF2122/97 ATMVPLLASGKSRKSPVIWWTVVTAAALGAMTPFILFAHGQVWQVGWIAG
Rv1635c|M.tuberculosis_H37Rv ATMVPLLASGKSRKSPVIWWTVVTAAALGAMTPFILFAHGQVWQVGWIAG
MLBr_01389|M.leprae_Br4923 AVILPWLAPNKFRNSPFIWWAVTSVVALGTITPFILFAHGQVWQVDWIFR
MAV_3138|M.avium_104 GVMLPLLTARGARRSAALWWAGSSAVAVGAMTPFLLFAHNQVWQVNWIYP
Mflv_0468|M.gilvum_PYR-GCK AAML---AATRPDRRTVRAWLVTVVVASVALIPLFVVLLSQQAQIAWIWP
MSMEG_0999|M.smegmatis_MC2_155 DSWITHVSGHPPGALLTFVWLDRIGLGGGAWAGLLCLLVGSSAAAAIVVA
: : * . : :. . .. :
MMAR_2439|M.marinum_M LNRNLFLDVVHRQYFDHS------------------------VAFAILAG
MUL_1618|M.ulcerans_Agy99 LNRNLFLDVVHRQYFDHS------------------------VAFAILAG
Mb1661c|M.bovis_AF2122/97 LNRNIILDVIHRQYFDHS------------------------VPFAILAG
Rv1635c|M.tuberculosis_H37Rv LNRNIILDVIHRQYFDHS------------------------VPFAILAG
MLBr_01389|M.leprae_Br4923 VSWHYVFDITQRQYFDHS------------------------VSFAIATA
MAV_3138|M.avium_104 VSWHYAFDIILRQYFDHS------------------------VALAVLSA
Mflv_0468|M.gilvum_PYR-GCK VGPVTLGQIAGEQYFPSVYSDSVRAVGPDQQQFSAEQLRVAMSAWARVAP
MSMEG_0999|M.smegmatis_MC2_155 LR-AVSGEATARLAAPFV--------------------------AVAPTA
: : . . :
MMAR_2439|M.marinum_M VLVVAAIAVRLTGIARP--------GEGTRPLLLASIAWVVIPTGIVLLY
MUL_1618|M.ulcerans_Agy99 VLVVAAIAVRLTGIARP--------GEGTRPLLLASIAWVVIPTGIVLLY
Mb1661c|M.bovis_AF2122/97 LIVAAGIAAHLAGARGP--------GGDTHRLVLVSAAWIVVPTAVVLIY
Rv1635c|M.tuberculosis_H37Rv LIVAAGIAAHLAGARGP--------GGDTHRLVLVSAAWIVVPTAVVLIY
MLBr_01389|M.leprae_Br4923 VIIVPAIATRLAGLRAP--------AGDLRSLVIICTAWIVIPTTLMVGY
MAV_3138|M.avium_104 VLIVAAAVARLAGVPAP--------PGDLRRLLILCAAWMVIPTALVVVY
Mflv_0468|M.gilvum_PYR-GCK LIVVIVIVAALAVRARRRHGALIENPSGTRVLVLGAVAWIVVPTVVLLGY
MSMEG_0999|M.smegmatis_MC2_155 IWIAVSADGYFAGVAAW----------GLALLALAVRDSVHRRRVALPTA
: :. :: . * : : :
MMAR_2439|M.marinum_M SAIVEPIYYPRYLILTAPAAAVILAVCIVTLAR-KPWLITGVVVLLAVAA
MUL_1618|M.ulcerans_Agy99 SAIVEPIYYPRYLILTAPAAAVILAVCIVTLAR-KPWLITGVVVLLAVAA
Mb1661c|M.bovis_AF2122/97 SATVEPIYYPRYLILTAPAAAVILAVCVVTIAR-KPWLIAGVVFLLAAAA
Rv1635c|M.tuberculosis_H37Rv SATVEPIYYPRYLILTAPAAAVILAVCVVTIAR-KPWLIAGVVFLLAAAA
MLBr_01389|M.leprae_Br4923 SAVIEPVYYPRYLILTAPAAAIVIAVCIVTVAR-KPWPIAGVLVLFAVAA
MAV_3138|M.avium_104 SAVAEPIYYPRYLIFTAPAMAIVLAVCIVTLAR-RPWPIAGAVLLCAVAA
Mflv_0468|M.gilvum_PYR-GCK SVLVRPLYQPHYLTFTTPALALLIGLCVVTLCRDDTRRLTATLAVLAVAA
MSMEG_0999|M.smegmatis_MC2_155 VGSGLLLGWGIFLNYGLALAALIALAVLITAPDWRAALRVAVPAVIAVLA
: :* . *:: ::* . ... : *. *
MMAR_2439|M.marinum_M FPNYFFTQRGPYAKEGWDYSQVADVISAHAAPGDCLLVDN-TVPWKPGPV
MUL_1618|M.ulcerans_Agy99 FPNYFFTQRGPYAKEGWDYSQVADVISTHAAPGDCLLVDN-TVPWKPGPV
Mb1661c|M.bovis_AF2122/97 FPNYFFTQRGPYAKEGWDYSQVADVISAHAKPGDCLLVDN-TAGWRPGPI
Rv1635c|M.tuberculosis_H37Rv FPNYFFTQRGPYAKEGWDYSQVADVISAHAKPGDCLLVDN-TAGWRPGPI
MLBr_01389|M.leprae_Br4923 FPNYLFTQRGRYAKEGWDYSQVADVISSQAAPGDCLIVDN-TVPWRPGPI
MAV_3138|M.avium_104 LPNYLFVQRWPYAKEGWDYSQVADLIGSHAAPGDCLLVDN-TVPWRPGPI
Mflv_0468|M.gilvum_PYR-GCK APNY-LAQRGPYAKYGSDYSQVAELLARESGPRDCVVVDDRTAPSVPHAI
MSMEG_0999|M.smegmatis_MC2_155 VVAVFLVAGFWWFDG---YTLVQERYWQGIANNRPFQYWS--WANLASVV
:. : . *: * : . . . :
MMAR_2439|M.marinum_M RALLATRPAAFRSLIDVERGAYGPKVGTLWDGHVAVWLTTEKINKCSTLW
MUL_1618|M.ulcerans_Agy99 RALLATRPAAFRSLIDVERGAYGPKVGTLWDGHVAVWLTTEKINKCSTLW
Mb1661c|M.bovis_AF2122/97 RALLATRPAAFRSLIDVERGTYGPKVGTLWDGHVAVWLTTAKIDKCPTLW
Rv1635c|M.tuberculosis_H37Rv RALLATRPAAFRSLIDVERGTYGPKVGTLWDGHVAVWLTTAKIDKCPTLW
MLBr_01389|M.leprae_Br4923 RALLAARPAAFRSLIDIERGFYGPTVGTLWDGHVPVWLVTAKINKCSTVW
MAV_3138|M.avium_104 RALLATRPAAFRSLIDVERGAYGPKVGTLWDGHVAIWLTTAKINKCSTIW
Mflv_0468|M.gilvum_PYR-GCK EGARRAHPVAAR-----DLGLAPPERTALFSARDPLGVRADDLRTCAVLW
MSMEG_0999|M.smegmatis_MC2_155 CAIGLGSVAGIGRVFDIKAARRNPGLPMLVIGALLAITFADLSMLSKAET
. .. . . * * . : . .
MMAR_2439|M.marinum_M TISNRDNALPDHEVGPALSPGTAFGKTPVYLFPHYLGFHIVERWQFHYSQ
MUL_1618|M.ulcerans_Agy99 TISNRDNALPDHEVGPALSPGTAFGKTPVYLFPHYLGFHIVERWQFHYSQ
Mb1661c|M.bovis_AF2122/97 TIANRDKSLPDHQVGEMLSPGTGFGRTPVYRFPSYLGFRIVERWQFHYSQ
Rv1635c|M.tuberculosis_H37Rv TIANRDKSLPDHQVGEMLSPGTGFGRTPVYRFPSYLGFRIVERWQFHYSQ
MLBr_01389|M.leprae_Br4923 TVSDRDTSLPDHQAGQLLSPGLILGRAPAYQFPSYLGFRIVERWQFHYSQ
MAV_3138|M.avium_104 TITNKDNSLPDHQSGQSLPPGSAFGQAPAYRFPGYLGFHIVERWQFHYSQ
Mflv_0468|M.gilvum_PYR-GCK TIG-------DAAPG---SP----ADAPVDRVARAQGFVPAERWRFNQTE
MSMEG_0999|M.smegmatis_MC2_155 ERIWLPFTVWLTAAGALLPP---------RSHRWWLGLNVAGALALNHLI
* .* *: . ::
MMAR_2439|M.marinum_M VIKSTR-----
MUL_1618|M.ulcerans_Agy99 VIKSTR-----
Mb1661c|M.bovis_AF2122/97 VVKSTR-----
Rv1635c|M.tuberculosis_H37Rv VVKSTR-----
MLBr_01389|M.leprae_Br4923 VIKSTR-----
MAV_3138|M.avium_104 VVKSTR-----
Mflv_0468|M.gilvum_PYR-GCK VVRSVPAGARS
MSMEG_0999|M.smegmatis_MC2_155 LTNW-------
: .