For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDEIQACCSATSLIASRESVGRDRVGAIERAGIDYLPEAARGSSPKNLFAVFVGGNLAFSTIIFGWLPIV MGLTFWQAVSSSVTGLLIGLMLTAPMSLFGPRTGTNNPVSSGAHFGVRGRLIGSSLTLMFALVYAAIAVW TSGEALIAGASRLFGAPQNHVAFLLGYGVIALEVILIALYGHATIVWAQKIIIPLAVVVLALGVLAYAPN FHPMGSAQHGYALQEFWPTWLFAVTVSVGGPLSYAPSVGDYTRRISRSRYSDRQIAVAVSAGIFLGLFST ATFGAFTASTFSAPTASYVADMVATAPGWYVVPLLILAVLGGCGQGVISIYSSGLDLESIVPRLNRTQTT LIASGIAVVLMYLGVVVTDAIDSVTGMTVVLNSFAGSWVAINICGFFKANRGQYDPKALQRFGQDGRGGL YWFSHGFNVRAVVPFIAGSGLGLLTVQNDLYQAPFANIAGGIDVSLLLSVVVGGGLYWLAQVSWPAPRVV SSGSSA
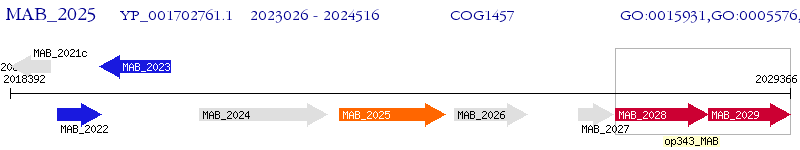
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2025 | - | - | 100% (496) | purine/cytosine permease |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0055 | - | 1e-27 | 23.35% (454) | permease for cytosine/purines, uracil, thiamine, allantoin |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4458 | - | 1e-87 | 36.12% (490) | transmembrane transport protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0160 | - | 1e-78 | 32.99% (479) | permease for cytosine/purines, uracil, thiamine, allantoin |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4458|M.smegmatis_MC2_155 -----------MATTEPVPVTP-RVTEVEQHGVEPIPDAERTARPLDLFR
Mvan_0160|M.vanbaalenii_PYR-1 -----------MAAETPALAPPPKVTEVETHGVEPIPDAERSARPLDLFR
MAB_2025|M.abscessus_ATCC_1997 MDEIQACCSATSLIASRESVGRDRVGAIERAGIDYLPEAARGSSPKNLFA
Mflv_0055|M.gilvum_PYR-GCK -------------------------MHDLEAELTVIPESAKTTDLRGQFW
: :*:: : : . *
MSMEG_4458|M.smegmatis_MC2_155 LVFGGANTIATVVLGTFPIIFGLSFRDALLATLAGLVLGALILAPMSLFG
Mvan_0160|M.vanbaalenii_PYR-1 LTFGGANTIATVVLGSFPIIFGLSFRDALLATLLGVVAGAAILAPMALFG
MAB_2025|M.abscessus_ATCC_1997 VFVGGNLAFSTIIFGWLPIVMGLTFWQAVSSSVTGLLIGLMLTAPMSLFG
Mflv_0055|M.gilvum_PYR-GCK IWAGANIAPINWILGALGVTMGLGLFETIAVLVVGNAIGMSLFGTFVVIG
: *. : . ::* : : :** : ::: : * * : ..: ::*
MSMEG_4458|M.smegmatis_MC2_155 PRNGTNNAVSSSAHLGVHGRVVGSFLSLLTAVAFFSISVWTSGDVLVGGA
Mvan_0160|M.vanbaalenii_PYR-1 PRNGTNNAVSSSAHLGVHGRVVGSFLSLLTAVAFFSISVWTSGDVLVGGA
MAB_2025|M.abscessus_ATCC_1997 PRTGTNNPVSSGAHFGVRGRLIGSSLTLMFALVYAAIAVWTSGEALIAGA
Mflv_0055|M.gilvum_PYR-GCK QKTGVTGMVLSRMVFGRRGAYVPAAVQAALCIGWCAVNTWIVLDLVMALL
:.*... * * :* :* : : : .: : :: .* : ::.
MSMEG_4458|M.smegmatis_MC2_155 NRAID----VPQNDIAVGVAYGIFAVLVLVVCIYGFRFMLLVNKIAVI--
Mvan_0160|M.vanbaalenii_PYR-1 TRAIGGAGSAAPNDWAVGVAYGVFALLVLTVCIYGFRFMLLVNKVAVV--
MAB_2025|M.abscessus_ATCC_1997 SRLFG----APQNHVAFLLGYGVIALEVILIALYGHATIVWAQKIIIP--
Mflv_0055|M.gilvum_PYR-GCK G-RIGLVDPELSNHAARIVVAGIIMTTQVVIAWFGYRLIAAFERWTVPPT
:. *. * : *:: : :. :*. : :: :
MSMEG_4458|M.smegmatis_MC2_155 AATLLFLAGVFAFGGVFDAGYAG-----SVALGDPLFWPSFVGAALIVMS
Mvan_0160|M.vanbaalenii_PYR-1 AATVLFLLGIFAFGGTFDPGYAGIFGPDADAATNALYWPSFVGAALIVMS
MAB_2025|M.abscessus_ATCC_1997 LAVVVLALGVLAYAPNFHPMGSA-----QHGYALQEFWPTWLFAVTVSVG
Mflv_0055|M.gilvum_PYR-GCK VAVLVAMTAVAWFGLDVDWGYTG----DAALTGAEHFTAITAVMTAIGIG
*.:: .: :. .. :. : . . : :.
MSMEG_4458|M.smegmatis_MC2_155 NPVSFGAFLGDWARYIPRDTPSWKPMLAAFSAQIATLVPFLFGLVTATVI
Mvan_0160|M.vanbaalenii_PYR-1 NPVSFGAFLGDWARYIPRDTPPWKPMAAAFCAQLATLVPFLFGLVTAAVV
MAB_2025|M.abscessus_ATCC_1997 GPLSYAPSVGDYTRRISRSRYSDRQIAVAVSAG-----IFLGLFSTATFG
Mflv_0055|M.gilvum_PYR-GCK WGITWLGYAGDYSRFVSSSVPARKLFAASALGQ------FIPVVWLGALG
::: **::* :. . . : : .: . *: . .:.
MSMEG_4458|M.smegmatis_MC2_155 ATTAPDYIA-NGDYVGGLLSVSPGWYFVPVCLIALIGGMSTGTTALYGTG
Mvan_0160|M.vanbaalenii_PYR-1 ATNAPQFID-EGNYVGGLLEVSPGWYFLPVCLIALIGGMSTGTTALYGTG
MAB_2025|M.abscessus_ATCC_1997 AFTASTFSAPTASYVADMVATAPGWYVVPLLILAVLGGCGQGVISIYSSG
Mflv_0055|M.gilvum_PYR-GCK ATLATLSAS---SDPGEIIVDAYGVLAIPVLLLVVHGPLATNILNIYTCA
* *. . . :: : * :*: ::.: * . . :* .
MSMEG_4458|M.smegmatis_MC2_155 LDFSSVFPKFSRVQATIFIGSIAIVFIFIGRFAFNVVQSISTFAVLIVTC
Mvan_0160|M.vanbaalenii_PYR-1 LDFSSVFPRFSRVQATVFIGVLAIVFIFIGRFAFNVVQSISTFAVLIVTC
MAB_2025|M.abscessus_ATCC_1997 LDLESIVPRLNRTQTTLIASGIAVVLMYLGVVVTDAIDSVTGMTVVLNSF
Mflv_0055|M.gilvum_PYR-GCK VSTQALDIHVNRRVLNIVIGVVAMAWVVYFVLNGDFAATIDAWLVGIVGW
:. .:: :..* .:. . :*:. : . : :: * :
MSMEG_4458|M.smegmatis_MC2_155 TAPWMVVMMIGWFTR-RGWYDSDALQVFNRRQRGGRYWFDHGWNWRGLTA
Mvan_0160|M.vanbaalenii_PYR-1 TAPWMVVMTTGWVVR-RGWYDSDSLQVFNRRQRGGRYWFARGWNFRGLGA
MAB_2025|M.abscessus_ATCC_1997 AGSWVAINICGFFKANRGQYDPKALQRFGQDGRGGLYWFSHGFNVRAVVP
Mflv_0055|M.gilvum_PYR-GCK LAPWAAVMLVHWFGVARRQVDPATLLTPPNES------TLPAVRPSALIS
..* .: :. * *. :* . . . .: .
MSMEG_4458|M.smegmatis_MC2_155 WLISAALS-ICFVNLPDQFVGPLGNLADGIDLSIPVGLGLAAVLYPALLW
Mvan_0160|M.vanbaalenii_PYR-1 WLVSAALS-LCFVNLPGQFVGPLGDLADGIDLSIPVGLGVAALLYPLLLV
MAB_2025|M.abscessus_ATCC_1997 FIAGSGLG-LLTVQN-DLYQAPFANIAGGIDVSLLLSVVVGGGLYWLAQV
Mflv_0055|M.gilvum_PYR-GCK LAVGIVFTWLFMYGMVPMMQGPIAVALGGVDLSWLAGGLAAGSLYAALEI
. : : .*:. .*:*:* . .. **
MSMEG_4458|M.smegmatis_MC2_155 LSPEPRDAFGPDGPRGVPSGAPANTPIESQD-TVISTHTEEVPA
Mvan_0160|M.vanbaalenii_PYR-1 AFPEPADAFGPEGPRFVRAGAPAGIPITSEDEAEKPVVSEGVTA
MAB_2025|M.abscessus_ATCC_1997 SWP---------APRVVSSGSSA---------------------
Mflv_0055|M.gilvum_PYR-GCK PKARRA--------------------------------------
.