For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSIMSVSALEQSAADVGDNSARQHAHGALPDSLAIAMLATVISGAWASRPSLWFDEAATISASASRTVPE LWRLLSHIDAVHGLYYLLMHGWFAIFPSTEFWSRVPSCLAIGAAAAGVTVFTRQFATRTTAVYAGIVFAI LPRITWAGIEARSSALSVAAAMWLTVLLVASVQRNRPRLWLCYALTLMLSILLNLTLATLVLVYAVILPW LAPNKFRNSPFIWWAVTSVVALGTITPFILFAHGQVWQVDWIFRVSWHYVFDITQRQYFDHSVSFAIATA VIIVPAIATRLAGLRAPAGDLRSLVIICTAWIVIPTTLMVGYSAVIEPVYYPRYLILTAPAAAIVIAVCI VTVARKPWPIAGVLVLFAVAAFPNYLFTQRGRYAKEGWDYSQVADVISSQAAPGDCLIVDNTVPWRPGPI RALLAARPAAFRSLIDIERGFYGPTVGTLWDGHVPVWLVTAKINKCSTVWTVSDRDTSLPDHQAGQLLSP GLILGRAPAYQFPSYLGFRIVERWQFHYSQVIKSTR*
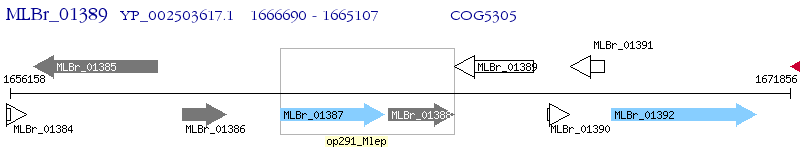
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_01389 | - | - | 100% (527) | hypothetical protein MLBr_01389 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1661c | - | 0.0 | 71.59% (528) | hypothetical protein Mb1661c |
| M. gilvum PYR-GCK | Mflv_0468 | - | 5e-89 | 36.83% (543) | hypothetical protein Mflv_0468 |
| M. tuberculosis H37Rv | Rv1635c | - | 0.0 | 71.59% (528) | transmembrane protein |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2439 | - | 0.0 | 70.40% (527) | transmembrane protein |
| M. avium 104 | MAV_3138 | - | 0.0 | 72.29% (498) | hypothetical protein MAV_3138 |
| M. smegmatis MC2 155 | MSMEG_3546 | - | 9e-05 | 23.38% (154) | hypothetical protein MSMEG_3546 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_1618 | - | 0.0 | 70.17% (523) | transmembrane protein |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1661c|M.bovis_AF2122/97 -----------MHASRPGAPPHAG----LPSRRTAGDQDHRADPKVTRIM
Rv1635c|M.tuberculosis_H37Rv -----------MHASRPGAPPHAG----LPSRRTAGDQDHRADPKVTRIM
MMAR_2439|M.marinum_M MEFQAAQHFSRLWAAQPGGPPKPRGQRPPNPRKMTRDQDHRWALTVTSIM
MUL_1618|M.ulcerans_Agy99 -------------------------------------------------M
MLBr_01389|M.leprae_Br4923 ---------------------------------------------MTSIM
MAV_3138|M.avium_104 --------------------------------------------------
Mflv_0468|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_3546|M.smegmatis_MC2_155 --------------------------------------------------
Mb1661c|M.bovis_AF2122/97 SASTLEQPAAAH-VDELVARMRGRLLDPLAIAVLAAVISGAWASRPSLWF
Rv1635c|M.tuberculosis_H37Rv SASTLEQPAAAH-VDELVARMRGRLLDPLAIAVLAAVISGAWASRPSLWF
MMAR_2439|M.marinum_M STPTLEQGATAT-DKAERVRPRGRRLDPWLMALLATLISVAWAGKPSLWF
MUL_1618|M.ulcerans_Agy99 STPTLEQGATAT-DKAERVRPRGRRLDPWLMALLATLISVAWAGRPSLWF
MLBr_01389|M.leprae_Br4923 SVSALEQSAADVGDNSARQHAHGALPDSLAIAMLATVISGAWASRPSLWF
MAV_3138|M.avium_104 ------------------------MLDPWAIAVLATALSAAWACRPSLWF
Mflv_0468|M.gilvum_PYR-GCK ---MLRGDGPGP--------ARQWSADAVVVGVLAFALGVAAAGRPSLWV
MSMEG_3546|M.smegmatis_MC2_155 -------------------------------------------MGKGYWF
. *.
Mb1661c|M.bovis_AF2122/97 DEGATISASASRTLPELWSLLGHIDAVHGLYYLLMHGWFAIFPPTELWSR
Rv1635c|M.tuberculosis_H37Rv DEGATISASASRTLPELWSLLGHIDAVHGLYYLLMHGWFAIFPPTELWSR
MMAR_2439|M.marinum_M DEGATISAAANRTLPELWKLLGHIDAVHGFYYLLMHGWYAIFPPTEFWSR
MUL_1618|M.ulcerans_Agy99 DEGATISAAANRTLPELWKLLGHIDAVHGFYYLLMHGWYAIFPPTEFWSR
MLBr_01389|M.leprae_Br4923 DEAATISASASRTVPELWRLLSHIDAVHGLYYLLMHGWFAIFPSTEFWSR
MAV_3138|M.avium_104 DEGATISAAASRTLPELWRLLGHIDAVHGAYYLLMHGWFALFPPTEFFSR
Mflv_0468|M.gilvum_PYR-GCK DEAGTLSAAT-RPVDALWTLVHHVDAVHGFYYLLLRGWFEIVPVTEFWAR
MSMEG_3546|M.smegmatis_MC2_155 DEVYMLAIGR---FHLDWGAADQPPVTPALAALADT----VAPGSLVTLR
** :: . . * : .. . * : * : . *
Mb1661c|M.bovis_AF2122/97 LPSCLAIGAAAAGVVVFAKQFSGRT-TAVCAGAVFAILPRVTWAGIEARS
Rv1635c|M.tuberculosis_H37Rv LPSCLAIGAAAAGVVVFAKQFSGRT-TAVCAGAVFAILPRVTWAGIEARS
MMAR_2439|M.marinum_M VPSSLAIGGAAAGVVVFTKQFSGRS-TALCAGAVFAILPRVTWAGIEARS
MUL_1618|M.ulcerans_Agy99 VPSSLAIGGAAAGVVVFTKQFSGRS-TALCAGAVFAILPRVTWAGIEARS
MLBr_01389|M.leprae_Br4923 VPSCLAIGAAAAGVTVFTRQFATRT-TAVYAGIVFAILPRITWAGIEARS
MAV_3138|M.avium_104 LPSALAVGAAAAGVTVFTRQFAPRR-TAVCAGAVFALLPRMTWAGMEARP
Mflv_0468|M.gilvum_PYR-GCK VPSALMVGVAAAGTCVLGSRLSGRK-VGIAAGVVFAVLPRTTWAAVEARP
MSMEG_3546|M.smegmatis_MC2_155 APAVLATGCAVVLAALIARELGGRRRAQAVTALAQATALFSTFAGHWLTP
*: * * *.. . :: .:. * . :. . * *:*. .
Mb1661c|M.bovis_AF2122/97 SALSVAAAVWLTVLLVAAVRCNTQRRWLLYALVLMLSILVSINLALLVPA
Rv1635c|M.tuberculosis_H37Rv SALSVAAAVWLTVLLVAAVRCNTQRRWLLYALVLMLSILVSINLALLVPA
MMAR_2439|M.marinum_M SALSVAAAVWLTVLLVAAVRRNRPWLWLLYALVLMVSILININLGLLVPV
MUL_1618|M.ulcerans_Agy99 SALSVAAAVWLTVLLVAAVRRDRPWLWLLYALVLMVSILININLGLLVPV
MLBr_01389|M.leprae_Br4923 SALSVAAAMWLTVLLVASVQRNRPRLWLCYALTLMLSILLNLTLATLVLV
MAV_3138|M.avium_104 YAFVAAAAVWLTVLFVAAVRRGAPRGWVGYALALMLAILLNLNMVLMVPV
Mflv_0468|M.gilvum_PYR-GCK YALSMACAVWLTVLFVAAVRRGGRRWWLGYGAALVGAVTVNVMVVLILAG
MSMEG_3546|M.smegmatis_MC2_155 YTLEPAQWLLLIWLLVRWIRTRDDRLLVAAGAATGLAAMTKFQVLLLCAV
:: * : * *:* :: : . . : .. : :
Mb1661c|M.bovis_AF2122/97 YATMVPLLASGKSRKSPVIWWTVVTAAALGAMTPFILFAHGQVWQVGWIA
Rv1635c|M.tuberculosis_H37Rv YATMVPLLASGKSRKSPVIWWTVVTAAALGAMTPFILFAHGQVWQVGWIA
MMAR_2439|M.marinum_M YGALLPLLAPKKSRKSPGIWWAVTSVVAIGAMTPFVLFAHGQVWQVGWIS
MUL_1618|M.ulcerans_Agy99 YGALLPLLAPKKSRKSPGIWWAVTSVVAIGAMTPFVLFAHGQVWQVGWIS
MLBr_01389|M.leprae_Br4923 YAVILPWLAPNKFRNSPFIWWAVTSVVALGTITPFILFAHGQVWQVDWIF
MAV_3138|M.avium_104 YGVMLPLLTARGARRSAALWWAGSSAVAVGAMTPFLLFAHNQVWQVNWIY
Mflv_0468|M.gilvum_PYR-GCK HAAML---AATRPDRRTVRAWLVTVVVASVALIPLFVVLLSQQAQIAWIW
MSMEG_3546|M.smegmatis_MC2_155 LLLGIAWCGPRALLRRPLLWVGAAVALAMVSPTLVWQHVHG--------W
: . . . . * : . .
Mb1661c|M.bovis_AF2122/97 GLNRNIILDVIHRQYFDHS------------------------VPFAILA
Rv1635c|M.tuberculosis_H37Rv GLNRNIILDVIHRQYFDHS------------------------VPFAILA
MMAR_2439|M.marinum_M GLNRNLFLDVVHRQYFDHS------------------------VAFAILA
MUL_1618|M.ulcerans_Agy99 GLNRNLFLDVVHRQYFDHS------------------------VAFAILA
MLBr_01389|M.leprae_Br4923 RVSWHYVFDITQRQYFDHS------------------------VSFAIAT
MAV_3138|M.avium_104 PVSWHYAFDIILRQYFDHS------------------------VALAVLS
Mflv_0468|M.gilvum_PYR-GCK PVGPVTLGQIAGEQYFPSVYSDSVRAVGPDQQQFSAEQLRVAMSAWARVA
MSMEG_3546|M.smegmatis_MC2_155 PQLQMTTVVAGEAEYLYGG-----------------------------RA
:*: :
Mb1661c|M.bovis_AF2122/97 GLIVAAGIAAHLAGARGP--------GGDTHRLVLVSAAWIVVPTAVVLI
Rv1635c|M.tuberculosis_H37Rv GLIVAAGIAAHLAGARGP--------GGDTHRLVLVSAAWIVVPTAVVLI
MMAR_2439|M.marinum_M GVLVVAAIAVRLTGIARP--------GEGTRPLLLASIAWVVIPTGIVLL
MUL_1618|M.ulcerans_Agy99 GVLVVAAIAVRLTGIARP--------GEGTRPLLLASIAWVVIPTGIVLL
MLBr_01389|M.leprae_Br4923 AVIIVPAIATRLAGLRAP--------AGDLRSLVIICTAWIVIPTTLMVG
MAV_3138|M.avium_104 AVLIVAAAVARLAGVPAP--------PGDLRRLLILCAAWMVIPTALVVV
Mflv_0468|M.gilvum_PYR-GCK PLIVVIVIVAALAVRARRRHGALIENPSGTRVLVLGAVAWIVVPTVVLLG
MSMEG_3546|M.smegmatis_MC2_155 GVAVQLILFAGVLGVGLG------VYGAWCLLRHAEFREYRFLPAVGVVL
: : . : : .:*: ::
Mb1661c|M.bovis_AF2122/97 YSATVEPIYYPRYLILTAPAAAVILAVCVVTIAR-KPWLIAGVVFLLAAA
Rv1635c|M.tuberculosis_H37Rv YSATVEPIYYPRYLILTAPAAAVILAVCVVTIAR-KPWLIAGVVFLLAAA
MMAR_2439|M.marinum_M YSAIVEPIYYPRYLILTAPAAAVILAVCIVTLAR-KPWLITGVVVLLAVA
MUL_1618|M.ulcerans_Agy99 YSAIVEPIYYPRYLILTAPAAAVILAVCIVTLAR-KPWLITGVVVLLAVA
MLBr_01389|M.leprae_Br4923 YSAVIEPVYYPRYLILTAPAAAIVIAVCIVTVAR-KPWPIAGVLVLFAVA
MAV_3138|M.avium_104 YSAVAEPIYYPRYLIFTAPAMAIVLAVCIVTLAR-RPWPIAGAVLLCAVA
Mflv_0468|M.gilvum_PYR-GCK YSVLVRPLYQPHYLTFTTPALALLIGLCVVTLCRDDTRRLTATLAVLAVA
MSMEG_3546|M.smegmatis_MC2_155 YVVFVATAGRPYYLGGLYAPFVAAGAVCLESVSPGTVRRWALRVGTVTAV
* . . * ** .. . .:*: ::. : : :..
Mb1661c|M.bovis_AF2122/97 AFPNYFFTQRGPYAKEGWDYSQVADVISAHAKPGDCLLVDN-TAGWRPGP
Rv1635c|M.tuberculosis_H37Rv AFPNYFFTQRGPYAKEGWDYSQVADVISAHAKPGDCLLVDN-TAGWRPGP
MMAR_2439|M.marinum_M AFPNYFFTQRGPYAKEGWDYSQVADVISAHAAPGDCLLVDN-TVPWKPGP
MUL_1618|M.ulcerans_Agy99 AFPNYFFTQRGPYAKEGWDYSQVADVISTHAAPGDCLLVDN-TVPWKPGP
MLBr_01389|M.leprae_Br4923 AFPNYLFTQRGRYAKEGWDYSQVADVISSQAAPGDCLIVDN-TVPWRPGP
MAV_3138|M.avium_104 ALPNYLFVQRWPYAKEGWDYSQVADLIGSHAAPGDCLLVDN-TVPWRPGP
Mflv_0468|M.gilvum_PYR-GCK AAPNY-LAQRGPYAKYGSDYSQVAELLARESGPRDCVVVDDRTAPSVPHA
MSMEG_3546|M.smegmatis_MC2_155 AATVAILVLSVNITPP-----DVGDRIAKAAATAYRDLPED-------ER
* . :. : :*.: :. : . : ::
Mb1661c|M.bovis_AF2122/97 IRALLATRPAAFRSLIDVERGTYGPKVGTLWDGHVAVWLTTAKIDKCPTL
Rv1635c|M.tuberculosis_H37Rv IRALLATRPAAFRSLIDVERGTYGPKVGTLWDGHVAVWLTTAKIDKCPTL
MMAR_2439|M.marinum_M VRALLATRPAAFRSLIDVERGAYGPKVGTLWDGHVAVWLTTEKINKCSTL
MUL_1618|M.ulcerans_Agy99 VRALLATRPAAFRSLIDVERGAYGPKVGTLWDGHVAVWLTTEKINKCSTL
MLBr_01389|M.leprae_Br4923 IRALLAARPAAFRSLIDIERGFYGPTVGTLWDGHVPVWLVTAKINKCSTV
MAV_3138|M.avium_104 IRALLATRPAAFRSLIDVERGAYGPKVGTLWDGHVAIWLTTAKINKCSTI
Mflv_0468|M.gilvum_PYR-GCK IEGARRAHPVAAR-----DLGLAPPERTALFSARDPLGVRADDLRTCAVL
MSMEG_3546|M.smegmatis_MC2_155 DRTVLLGESYITAAYLDGAAPRYG--LPQAYGTNRSYGYFTPPPDTADEA
. .. :. . . : ..
Mb1661c|M.bovis_AF2122/97 WTIANRDKSLPDHQVGEMLSPGTGFGRTPVYRFPSYLGFRIVERWQFHYS
Rv1635c|M.tuberculosis_H37Rv WTIANRDKSLPDHQVGEMLSPGTGFGRTPVYRFPSYLGFRIVERWQFHYS
MMAR_2439|M.marinum_M WTISNRDNALPDHEVGPALSPGTAFGKTPVYLFPHYLGFHIVERWQFHYS
MUL_1618|M.ulcerans_Agy99 WTISNRDNALPDHEVGPALSPGTAFGKTPVYLFPHYLGFHIVERWQFHYS
MLBr_01389|M.leprae_Br4923 WTVSDRDTSLPDHQAGQLLSPGLILGRAPAYQFPSYLGFRIVERWQFHYS
MAV_3138|M.avium_104 WTITNKDNSLPDHQSGQSLPPGSAFGQAPAYRFPGYLGFHIVERWQFHYS
Mflv_0468|M.gilvum_PYR-GCK WTIG-------DAAPG---SP----ADAPVDRVARAQGFVPAERWRFNQT
MSMEG_3546|M.smegmatis_MC2_155 LYVGAEPDGMREHFVT----------VRNLATIDGDLKLFLLRERRHPWP
: : . : .. :. .
Mb1661c|M.bovis_AF2122/97 QVVKSTR-----
Rv1635c|M.tuberculosis_H37Rv QVVKSTR-----
MMAR_2439|M.marinum_M QVIKSTR-----
MUL_1618|M.ulcerans_Agy99 QVIKSTR-----
MLBr_01389|M.leprae_Br4923 QVIKSTR-----
MAV_3138|M.avium_104 QVVKSTR-----
Mflv_0468|M.gilvum_PYR-GCK EVVRSVPAGARS
MSMEG_3546|M.smegmatis_MC2_155 QIWSEQRSLTVS
:: .