For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTSQTGVRDELLHAGVRLLDDHGPDALQTRKVAAAAGTSTMAVYTHFGGMRGLIAAIAEEGLRQFDVALT VPQTADPVADLLAIGTAYRRYAIERPHMYRLMFGSTSAHGINVPARDVLTLKVAEIEHQHPSFAHVVRAV HRCLLAGRFATALGADDDTAIVATAAQFWSQIHGFVMLELAGFYGDRGAAVEPVLAAMTVNLLVALGDSP ERAQCSLRAEQTQKNTLGRAT
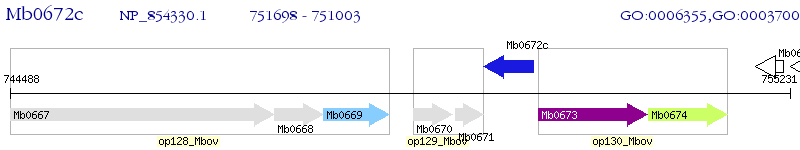
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0672c | - | - | 100% (231) | TetR family transcriptional regulator |
| M. bovis AF2122 / 97 | Mb0700 | - | 3e-07 | 25.14% (183) | TetR family transcriptional regulator |
| M. bovis AF2122 / 97 | Mb1846 | - | 9e-06 | 22.81% (228) | transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2110 | - | 1e-38 | 42.99% (214) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0653c | - | 1e-129 | 100.00% (231) | TetR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3875 | - | 1e-63 | 58.41% (214) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0992 | - | 6e-88 | 71.00% (231) | transcriptional regulatory protein |
| M. avium 104 | MAV_4506 | - | 3e-87 | 75.46% (216) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_1397 | - | 1e-07 | 24.87% (189) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_4347 | - | 4e-42 | 46.95% (213) | PUTATIVE transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_0743 | - | 2e-87 | 70.69% (232) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_5135 | - | 2e-10 | 29.26% (188) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2110|M.gilvum_PYR-GCK ------------------------MVSTVRD---NLVAAGVRLLEGGGVS
TH_4347|M.thermoresistible__bu -----------------------VTADDIRE---KLVDAGVRLLERDGVQ
Mb0672c|M.bovis_AF2122/97 ------------------MTS----QTGVRD---ELLHAGVRLLDDHGPD
Rv0653c|M.tuberculosis_H37Rv ------------------MTS----QTGVRD---ELLHAGVRLLDDHGPD
MMAR_0992|M.marinum_M ---------------MIGETTSPTQRSGVRD---EMLHAAVALLDAHGPD
MUL_0743|M.ulcerans_Agy99 ---------------MIGETTSPTQRSGVRD---EMLHAAVALLDAHGPD
MAV_4506|M.avium_104 ------------------MTSQPAPQRNVRD---GMLAAAVALLHEHGAD
MAB_3875|M.abscessus_ATCC_1997 MNVLPLCEHGKTMSSSARRNSEGTDRAVVRNPRVDLVSAAVRLLNEQGPD
Mvan_5135|M.vanbaalenii_PYR-1 -----------------MSSRGTYHHGDLRA---TILARAADLVAERGAD
MSMEG_1397|M.smegmatis_MC2_155 -------------------MRATRTTRLSRD---TIVNAALTFLDREGWD
* :: . :: * .
Mflv_2110|M.gilvum_PYR-GCK ALSARSVATEAGTSTMALYTHFGGMTGLLDAVAVDVFARFTEALTEVAPT
TH_4347|M.thermoresistible__bu ALSARNLAAEAGTSTMAVYTRFGGMPGVLEAVAGEIFARFADALTQIPQT
Mb0672c|M.bovis_AF2122/97 ALQTRKVAAAAGTSTMAVYTHFGGMRGLIAAIAEEGLRQFDVALTVP-QT
Rv0653c|M.tuberculosis_H37Rv ALQTRKVAAAAGTSTMAVYTHFGGMRGLIAAIAEEGLRQFDVALTVP-QT
MMAR_0992|M.marinum_M ALQTRKVAGAAGTSTMAVYTHFGGMPELIAEVAEEGLRQFDTALAVP-PS
MUL_0743|M.ulcerans_Agy99 ALQTRKVAGATGTSTMAVYTHFGGMPELIAEVADEGLRQFDTALAVP-PS
MAV_4506|M.avium_104 ALQTRKVAGAAGTSTMAVYTHFGGMRGLIAEVAEEGLRQFDAALTVP-PT
MAB_3875|M.abscessus_ATCC_1997 ALQTRKIAAAAGTSTMAVYTYFGGMRELIAEVAEAGLRQFAEAQAAVEQS
Mvan_5135|M.vanbaalenii_PYR-1 GVSLRELARAAGVSHAAPAHHFTDRRGLFTALATEGFRSLAEELSRA---
MSMEG_1397|M.smegmatis_MC2_155 ALTINALANQLGTKGPSLYNHVHSLDDLRRTVRMRVIDDIISMLNRVGEG
.: . :* *.. : . . : : : :
Mflv_2110|M.gilvum_PYR-GCK DDPVADFLMMGLAYHRFALANPQRYQLIFGAAAPASISRFRADITETGSA
TH_4347|M.thermoresistible__bu GDPVADFLLMGAAYREYALTHPQRYLLMFGTATPASIAGSRADLTATGEV
Mb0672c|M.bovis_AF2122/97 ADPVADLLAIGTAYRRYAIERPHMYRLMFGSTSAHGIN-VPARDVLTLKV
Rv0653c|M.tuberculosis_H37Rv ADPVADLLAIGTAYRRYAIERPHMYRLMFGSTSAHGIN-VPARDVLTLKV
MMAR_0992|M.marinum_M DDPVADLVATGAAYRRYAIERPHMYRLMFGSTSAHGIN-APTQNVLTLTI
MUL_0743|M.ulcerans_Agy99 DDPVADLVATGAAYRRYAIERPHMYRLMFGSTSAHGIN-APAQNVLTLTI
MAV_4506|M.avium_104 DDPVADLFVIGAAYRRYAIKHRHMYRLMFGSTSAHGIN-APAHNVLTLTV
MAB_3875|M.abscessus_ATCC_1997 DDVIADFMLTGMAYRQFAIDYPHMYRLMFGVTSAHGVN-APRRNMFDT--
Mvan_5135|M.vanbaalenii_PYR-1 ---RPEFLDAALAYVRFALANPGHYEVMFDKSLYHADD--------PALV
MSMEG_1397|M.smegmatis_MC2_155 RTRDDAVIAMASAYRSYAHHHPGRYSAFTRMPFGDDDP------------
.. . ** :* * : .
Mflv_2110|M.gilvum_PYR-GCK TNRAEWAASFGALHGAVHRMMAAGRIR---------------------RD
TH_4347|M.thermoresistible__bu SDRTEWAASFEALRTMVRRMIAAGRIR---------------------DD
Mb0672c|M.bovis_AF2122/97 AEIEHQHPSFAHVVRAVHRCLLAGRFATALGADD--------------DT
Rv0653c|M.tuberculosis_H37Rv AEIEHQHPSFAHVVRAVHRCLLAGRFATALGADD--------------DT
MMAR_0992|M.marinum_M SQIEQRHPSFAHVVRGVRRSMLAGRITVNGGADRNRASAPDGLATDVPDA
MUL_0743|M.ulcerans_Agy99 SQIEQRHPSFAHVVRGVRRSMLAGRITVNGGDDRNRAAAPDGLATDVLDA
MAV_4506|M.avium_104 AEIEQNYPSFAHVVRAVHRCMRAGRITAGSADD---------------DG
MAB_3875|M.abscessus_ATCC_1997 SHVTPEHPSAAYLFRCVQRAMAAGRIDGSPED----------------AA
Mvan_5135|M.vanbaalenii_PYR-1 DAETAAGSELAAGVGTLDDPRAK--------------------------D
MSMEG_1397|M.smegmatis_MC2_155 EYSAATKAAAAPVIEVLASFGLEG-------------------------E
. :
Mflv_2110|M.gilvum_PYR-GCK DELAVAGRLWSINHGAVMLEMAGFFGHEGHGLVHILGPLIIDTLVGMGDD
TH_4347|M.thermoresistible__bu GEAAIAGRLWSLIHGEVMLELAGFFGAEGHGLTKILTPMTVDMLVGLGDD
Mb0672c|M.bovis_AF2122/97 AIVATAAQFWSQIHGFVMLELAGFYGDRGAAVEPVLAAMTVNLLVALGDS
Rv0653c|M.tuberculosis_H37Rv AIVATAAQFWSQIHGFVMLELAGFYGDRGAAVEPVLAAMTVNLLVALGDS
MMAR_0992|M.marinum_M EIVATAAQFWALIHGFVMLELAGFFGDEGAAIEPVLASMTTNLLVALGDS
MUL_0743|M.ulcerans_Agy99 EIVATAAQFWALIHGFVMLELAGFFGDEGAAIEPVLASMTTNLLVALGDS
MAV_4506|M.avium_104 AVVATAAQFWASIHGFVMLELAGYCGDDGSAVLPVLGSMTSNLLVALGDS
MAB_3875|M.abscessus_ATCC_1997 KVAATAAKFWTVMHGFVMLELAGFWGEDGAAVGPVLASMTTDLLVALGDT
Mvan_5135|M.vanbaalenii_PYR-1 DPQAAALAAWSLVHGFALLWLNGAVNTTG----DPMATVRRVAGMLFGGE
MSMEG_1397|M.smegmatis_MC2_155 QAFYAALEFWSALHGFVLLEMTGVMDDVDT------DAVFTDMLLRLASG
* *: ** .:* : * . . .: : :..
Mflv_2110|M.gilvum_PYR-GCK RDAATLSMQTVVARLG------
TH_4347|M.thermoresistible__bu RARVEASLTTAAEAHGR-----
Mb0672c|M.bovis_AF2122/97 PERAQCSLRAEQTQKNTLGRAT
Rv0653c|M.tuberculosis_H37Rv PERAQCSLRAEQTQKNTLGRAT
MMAR_0992|M.marinum_M PELAKRSQRLATGAG-------
MUL_0743|M.ulcerans_Agy99 PELAKRSQRLTTGAG-------
MAV_4506|M.avium_104 PKRVSQSMRSAMLGS-------
MAB_3875|M.abscessus_ATCC_1997 PERLNSSAAAAAARLAPSA---
Mvan_5135|M.vanbaalenii_PYR-1 PVTCPT----------------
MSMEG_1397|M.smegmatis_MC2_155 MERREA----------------