For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IGETTSPTQRSGVRDEMLHAAVALLDAHGPDALQTRKVAGATGTSTMAVYTHFGGMPELIAEVADEGLRQ FDTALAVPPSDDPVADLVATGAAYRRYAIERPHMYRLMFGSTSAHGINAPAQNVLTLTISQIEQRHPSFA HVVRGVRRSMLAGRITVNGGDDRNRAAAPDGLATDVLDAEIVATAAQFWALIHGFVMLELAGFFGDEGAA IEPVLASMTTNLLVALGDSPELAKRSQRLTTGAG*
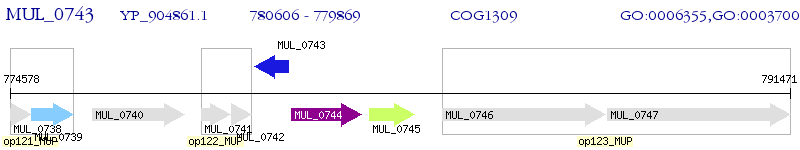
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0743 | - | - | 100% (245) | transcriptional regulatory protein |
| M. ulcerans Agy99 | MUL_0990 | - | 8e-06 | 25.33% (229) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0672c | - | 2e-87 | 70.69% (232) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2110 | - | 6e-33 | 37.83% (230) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0653c | - | 2e-87 | 70.69% (232) | TetR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3875 | - | 8e-65 | 57.89% (228) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0992 | - | 1e-133 | 97.14% (245) | transcriptional regulatory protein |
| M. avium 104 | MAV_4506 | - | 1e-86 | 69.36% (235) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_1397 | - | 3e-07 | 25.98% (204) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_4347 | - | 6e-37 | 41.20% (233) | PUTATIVE transcriptional regulator, TetR family |
| M. vanbaalenii PYR-1 | Mvan_3167 | - | 1e-07 | 25.55% (227) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2110|M.gilvum_PYR-GCK ------------------------MVSTVRD----NLVAAGVRLLEGGGV
TH_4347|M.thermoresistible__bu -----------------------VTADDIRE----KLVDAGVRLLERDGV
Mb0672c|M.bovis_AF2122/97 ------------------MTS----QTGVRD----ELLHAGVRLLDDHGP
Rv0653c|M.tuberculosis_H37Rv ------------------MTS----QTGVRD----ELLHAGVRLLDDHGP
MUL_0743|M.ulcerans_Agy99 ---------------MIGETTSPTQRSGVRD----EMLHAAVALLDAHGP
MMAR_0992|M.marinum_M ---------------MIGETTSPTQRSGVRD----EMLHAAVALLDAHGP
MAV_4506|M.avium_104 ------------------MTSQPAPQRNVRD----GMLAAAVALLHEHGA
MAB_3875|M.abscessus_ATCC_1997 MNVLPLCEHGKTMSSSARRNSEGTDRAVVRNPRV-DLVSAAVRLLNEQGP
MSMEG_1397|M.smegmatis_MC2_155 -------------------MRATRTTRLSRD----TIVNAALTFLDREGW
Mvan_3167|M.vanbaalenii_PYR-1 ----------------MCDTGRVGKRQDTRDRIERQIVEIGRAHLVTHGA
*: :: . * *
Mflv_2110|M.gilvum_PYR-GCK SALSARSVATEAGTSTMALYTHFGGMTGLLDAVAVDVFARFTEALTEVA-
TH_4347|M.thermoresistible__bu QALSARNLAAEAGTSTMAVYTRFGGMPGVLEAVAGEIFARFADALTQIP-
Mb0672c|M.bovis_AF2122/97 DALQTRKVAAAAGTSTMAVYTHFGGMRGLIAAIAEEGLRQFDVALTVP--
Rv0653c|M.tuberculosis_H37Rv DALQTRKVAAAAGTSTMAVYTHFGGMRGLIAAIAEEGLRQFDVALTVP--
MUL_0743|M.ulcerans_Agy99 DALQTRKVAGATGTSTMAVYTHFGGMPELIAEVADEGLRQFDTALAVP--
MMAR_0992|M.marinum_M DALQTRKVAGAAGTSTMAVYTHFGGMPELIAEVAEEGLRQFDTALAVP--
MAV_4506|M.avium_104 DALQTRKVAGAAGTSTMAVYTHFGGMRGLIAEVAEEGLRQFDAALTVP--
MAB_3875|M.abscessus_ATCC_1997 DALQTRKIAAAAGTSTMAVYTYFGGMRELIAEVAEAGLRQFAEAQAAVE-
MSMEG_1397|M.smegmatis_MC2_155 DALTINALANQLGTKGPSLYNHVHSLDDLRRTVRMRVIDDIISMLNRVG-
Mvan_3167|M.vanbaalenii_PYR-1 AALSLRAIARELGMVSSAVYRYVASRDELLTLLLVDAYGELADSVERARD
** . :* * ::* . . : : :
Mflv_2110|M.gilvum_PYR-GCK -PTDDPVADFLMMGLAYHRFALANPQRYQLIFGAAAPASISRFRADITET
TH_4347|M.thermoresistible__bu -QTGDPVADFLLMGAAYREYALTHPQRYLLMFGTATPASIAGSRADLTAT
Mb0672c|M.bovis_AF2122/97 -QTADPVADLLAIGTAYRRYAIERPHMYRLMFGSTSAHGIN-VPARDVLT
Rv0653c|M.tuberculosis_H37Rv -QTADPVADLLAIGTAYRRYAIERPHMYRLMFGSTSAHGIN-VPARDVLT
MUL_0743|M.ulcerans_Agy99 -PSDDPVADLVATGAAYRRYAIERPHMYRLMFGSTSAHGIN-APAQNVLT
MMAR_0992|M.marinum_M -PSDDPVADLVATGAAYRRYAIERPHMYRLMFGSTSAHGIN-APTQNVLT
MAV_4506|M.avium_104 -PTDDPVADLFVIGAAYRRYAIKHRHMYRLMFGSTSAHGIN-APAHNVLT
MAB_3875|M.abscessus_ATCC_1997 -QSDDVIADFMLTGMAYRQFAIDYPHMYRLMFGVTSAHGVN-APRRNMFD
MSMEG_1397|M.smegmatis_MC2_155 -EGRTRDDAVIAMASAYRSYAHHHPGRYSAFTRMPFGDDDP---------
Mvan_3167|M.vanbaalenii_PYR-1 AAAGDWQDRLRAVAHATRAWAVHHPARWALLYGSPVPGYHAPREQTVGPG
. . * : :* : : .
Mflv_2110|M.gilvum_PYR-GCK GSATNRAEWAASFGALHGAVHRMMAAGRIR--------------------
TH_4347|M.thermoresistible__bu GEVSDRTEWAASFEALRTMVRRMIAAGRIR--------------------
Mb0672c|M.bovis_AF2122/97 LKVAEIEHQHPSFAHVVRAVHRCLLAGRFATALGADD-------------
Rv0653c|M.tuberculosis_H37Rv LKVAEIEHQHPSFAHVVRAVHRCLLAGRFATALGADD-------------
MUL_0743|M.ulcerans_Agy99 LTISQIEQRHPSFAHVVRGVRRSMLAGRITVNGGDDRNRAAAPDGLATDV
MMAR_0992|M.marinum_M LTISQIEQRHPSFAHVVRGVRRSMLAGRITVNGGADRNRASAPDGLATDV
MAV_4506|M.avium_104 LTVAEIEQNYPSFAHVVRAVHRCMRAGRITAGSADD--------------
MAB_3875|M.abscessus_ATCC_1997 T--SHVTPEHPSAAYLFRCVQRAMAAGRIDGSPED---------------
MSMEG_1397|M.smegmatis_MC2_155 -------EYSAATKAAAAPVIEVLASFGLE--------------------
Mvan_3167|M.vanbaalenii_PYR-1 TRVVGLLFSAIAEGVAAGAVRTSKVTVPQSLSSDFDRLR-----AEFEFV
: * :
Mflv_2110|M.gilvum_PYR-GCK -RDDELAVAGRLWSINHGAVMLEMAGFFGHEGHGLVHILGPLIIDTLVGM
TH_4347|M.thermoresistible__bu -DDGEAAIAGRLWSLIHGEVMLELAGFFGAEGHGLTKILTPMTVDMLVGL
Mb0672c|M.bovis_AF2122/97 -DTAIVATAAQFWSQIHGFVMLELAGFYGDRGAAVEPVLAAMTVNLLVAL
Rv0653c|M.tuberculosis_H37Rv -DTAIVATAAQFWSQIHGFVMLELAGFYGDRGAAVEPVLAAMTVNLLVAL
MUL_0743|M.ulcerans_Agy99 LDAEIVATAAQFWALIHGFVMLELAGFFGDEGAAIEPVLASMTTNLLVAL
MMAR_0992|M.marinum_M PDAEIVATAAQFWALIHGFVMLELAGFFGDEGAAIEPVLASMTTNLLVAL
MAV_4506|M.avium_104 -DGAVVATAAQFWASIHGFVMLELAGYCGDDGSAVLPVLGSMTSNLLVAL
MAB_3875|M.abscessus_ATCC_1997 -AAKVAATAAKFWTVMHGFVMLELAGFWGEDGAAVGPVLASMTTDLLVAL
MSMEG_1397|M.smegmatis_MC2_155 -GEQAFYAALEFWSALHGFVLLEMTGVMDDVD------TDAVFTDMLLRL
Mvan_3167|M.vanbaalenii_PYR-1 ADDDVVARCLVLWAALVGAISLEVFGQYGADTFADASVVFDAQMDLVIDL
. :*: * : **: * . : :: :
Mflv_2110|M.gilvum_PYR-GCK GDDRDAATLSMQTVVARLG------
TH_4347|M.thermoresistible__bu GDDRARVEASLTTAAEAHGR-----
Mb0672c|M.bovis_AF2122/97 GDSPERAQCSLRAEQTQKNTLGRAT
Rv0653c|M.tuberculosis_H37Rv GDSPERAQCSLRAEQTQKNTLGRAT
MUL_0743|M.ulcerans_Agy99 GDSPELAKRSQRLTTGAG-------
MMAR_0992|M.marinum_M GDSPELAKRSQRLATGAG-------
MAV_4506|M.avium_104 GDSPKRVSQSMRSAMLGS-------
MAB_3875|M.abscessus_ATCC_1997 GDTPERLNSSAAAAAARLAPSA---
MSMEG_1397|M.smegmatis_MC2_155 ASGMERREA----------------
Mvan_3167|M.vanbaalenii_PYR-1 FCRQGAN------------------