For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SSRGTYHHGDLRATILARAADLVAERGADGVSLRELARAAGVSHAAPAHHFTDRRGLFTALATEGFRSLA EELSRARPEFLDAALAYVRFALANPGHYEVMFDKSLYHADDPALVDAETAAGSELAAGVGTLDDPRAKDD PQAAALAAWSLVHGFALLWLNGAVNTTGDPMATVRRVAGMLFGGEPVTCPT*
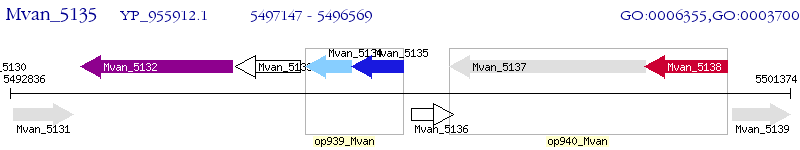
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5135 | - | - | 100% (192) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0895 | - | 3e-14 | 29.05% (179) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_4572 | - | 4e-11 | 32.72% (162) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0672c | - | 1e-10 | 29.26% (188) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1618 | - | 3e-77 | 75.27% (186) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0653c | - | 1e-10 | 29.26% (188) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_00717 | - | 2e-06 | 32.26% (93) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_0499 | - | 3e-51 | 56.91% (181) | TetR family transcriptional regulator |
| M. marinum M | MMAR_4906 | - | 4e-64 | 67.39% (184) | transcriptional regulator |
| M. avium 104 | MAV_0728 | - | 2e-65 | 68.28% (186) | TetR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_5838 | - | 4e-72 | 72.63% (179) | TetR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_4347 | - | 2e-08 | 27.72% (184) | PUTATIVE transcriptional regulator, TetR family |
| M. ulcerans Agy99 | MUL_0500 | - | 7e-64 | 65.45% (191) | transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0672c|M.bovis_AF2122/97 ------MTSQTGVRDELLHAGVRLLDDHGPDALQTRKVAAAAGTSTMAVY
Rv0653c|M.tuberculosis_H37Rv ------MTSQTGVRDELLHAGVRLLDDHGPDALQTRKVAAAAGTSTMAVY
TH_4347|M.thermoresistible__bu -------VTADDIREKLVDAGVRLLERDGVQALSARNLAAEAGTSTMAVY
Mvan_5135|M.vanbaalenii_PYR-1 -MSSRGTYHHGDLRATILARAADLVAERGADGVSLRELARAAGVSHAAPA
Mflv_1618|M.gilvum_PYR-GCK --MTRGSYHHGDLKAVILAQAATLVAERGADGVSLRELARAAGVSHAAPA
MSMEG_5838|M.smegmatis_MC2_155 -----MAYHHGDLKAVILQHAATLVAERGADGISLRELARTAGVSHAAPA
MMAR_4906|M.marinum_M --MNRPAYHHGDLRAVILDEAARLVAQRGADRVSMRELARDAGVSHAAPA
MUL_0500|M.ulcerans_Agy99 --MNRPAYHHGDLRAVILDEAARLVAQRGADRVSMRELARDAGVSHAAPA
MAV_0728|M.avium_104 --MTRHSYHHGDLPAVILAEAARLVAERGAERVSLRELARCAGVSHAAPA
MAB_0499|M.abscessus_ATCC_1997 -----MSYHHGDLRAAILTSAADMVARRGATELSLRELAREAGVSHAAPA
MLBr_00717|M.leprae_Br4923 MTTARRRLSPEDRRAELLALGAEVFGKRPYDEVRIDEIAERAGVSRALMY
. :: .. :. : ::* **.*
Mb0672c|M.bovis_AF2122/97 THFGGMRGLIAAIAEEGLRQFDVALT-VPQTADPVADLLAIGTAYRRYAI
Rv0653c|M.tuberculosis_H37Rv THFGGMRGLIAAIAEEGLRQFDVALT-VPQTADPVADLLAIGTAYRRYAI
TH_4347|M.thermoresistible__bu TRFGGMPGVLEAVAGEIFARFADALTQIPQTGDPVADFLLMGAAYREYAL
Mvan_5135|M.vanbaalenii_PYR-1 HHFTDRRGLFTALATEGFRSLAEELS------RARPEFLDAALAYVRFAL
Mflv_1618|M.gilvum_PYR-GCK HHFTDRRGVFTALATQGWRMLAAALG------ESRPEFLDAALAYVRFAV
MSMEG_5838|M.smegmatis_MC2_155 HHFTDRRGLFTALATEGYQLLADALK------DVRGQFIDAAKAYVRFAL
MMAR_4906|M.marinum_M HHFTDRRGLFTALATQGFQLLAAALV------AARGHFADAALAYVRFAI
MUL_0500|M.ulcerans_Agy99 HHFTDRRGLFTALATQGFQLLAAALV------AARGHFADAALAYVRFAI
MAV_0728|M.avium_104 HHFTDRRGLFTALATQGFELLTRALA------DARGDFADAALAYVQFAL
MAB_0499|M.abscessus_ATCC_1997 HHFGDRRGLFTALAADGFTRLADALD------GAHPDFHAAALAYVEFAL
MLBr_00717|M.leprae_Br4923 HYFPDKRAFFAAVVKDEADRLYAATN--IPPPAGLTMFEEVRVGVLAYMA
* . ..: *:. : : : . :
Mb0672c|M.bovis_AF2122/97 ERPHMYRLMFGSTS-AHGIN-VPARDVLTLKVAEIEHQHPSFAHVVRAVH
Rv0653c|M.tuberculosis_H37Rv ERPHMYRLMFGSTS-AHGIN-VPARDVLTLKVAEIEHQHPSFAHVVRAVH
TH_4347|M.thermoresistible__bu THPQRYLLMFGTAT-PASIAGSRADLTATGEVSDRTEWAASFEALRTMVR
Mvan_5135|M.vanbaalenii_PYR-1 ANPGHYEVMFDKSL-YHADD----PALVDAETAAGSELAAGVGTLDDPRA
Mflv_1618|M.gilvum_PYR-GCK DHPGHYEVMFDRSL-VDPDN----PELVAAQTEAGAELAAGVGTLDDARA
MSMEG_5838|M.smegmatis_MC2_155 DHPGHYAVMFDKSL-YDDTD----PGLVAAASAAGAELNRGVGTLDDPKA
MMAR_4906|M.marinum_M EHPGHYQVMFNRSL-HDADD----ADLAAAQAAAAAELAAGVATLRDPGA
MUL_0500|M.ulcerans_Agy99 EHPGHYQVMFNRSL-HDADD----ADLAAAQAAAAAELAAGVATLRDPGA
MAV_0728|M.avium_104 DHPGHYQVMFNRSL-LDASD----GGLAAAEGAAGAELSRGVATLRDPHA
MAB_0499|M.abscessus_ATCC_1997 ANPGHYSVMFEPAL-LDLAD----GRLAEARDRAAAALDSGIATLTPQQT
MLBr_00717|M.leprae_Br4923 YHKQNPEAAWAAYVGLGRSDPVLLGIDDEAKNRQMEHIMSRIGDVVSVVP
. : . :
Mb0672c|M.bovis_AF2122/97 RCLLAGRFATALGADDDTAIVATAAQFWSQIHGFVMLELAGFYGDRGAAV
Rv0653c|M.tuberculosis_H37Rv RCLLAGRFATALGADDDTAIVATAAQFWSQIHGFVMLELAGFYGDRGAAV
TH_4347|M.thermoresistible__bu RMIAAGRIR-------DDGEAAIAGRLWSLIHGEVMLELAGFFGAEGHGL
Mvan_5135|M.vanbaalenii_PYR-1 KDDPQ----------------AAALAAWSLVHGFALLWLNGAVNTTG---
Mflv_1618|M.gilvum_PYR-GCK REDPQ----------------AAGLAAWSLVHGFAMLWLNEAIDTDA---
MSMEG_5838|M.smegmatis_MC2_155 KDDPA----------------GAALAAWSMVHGFSLLWLNDAIDTDR---
MMAR_4906|M.marinum_M RSDPE----------------GAQLAAWSLVHGFSTLWLNDALDAQVRAL
MUL_0500|M.ulcerans_Agy99 RFDPE----------------GAQLAAWSLVHGFSTLWLNDAFDAQVRAL
MAV_0728|M.avium_104 RADPG----------------GAELAAWSLVHGFATLWLNDAVDAAVRAA
MAB_0499|M.abscessus_ATCC_1997 SADRI----------------TAARAAWSLVHGFVSLWTTGALADSRDDA
MLBr_00717|M.leprae_Br4923 ESKLHP---------------DIERDLRVIVHGWLAFTFEVCRQRIMDPS
:** :
Mb0672c|M.bovis_AF2122/97 EPVLAAMTVNLLVALGDSPERAQCSLRAEQTQKNTLGRAT----------
Rv0653c|M.tuberculosis_H37Rv EPVLAAMTVNLLVALGDSPERAQCSLRAEQTQKNTLGRAT----------
TH_4347|M.thermoresistible__bu TKILTPMTVDMLVGLGDDRARVEASL---TTAAEAHGR------------
Mvan_5135|M.vanbaalenii_PYR-1 DPMATVRRVAGMLFGGEPVTCPT---------------------------
Mflv_1618|M.gilvum_PYR-GCK DAVVTARRVARMLFTPGPVGDAGRG-------------------------
MSMEG_5838|M.smegmatis_MC2_155 DPIATVERLGAILFDG----------------------------------
MMAR_4906|M.marinum_M DPMDTVLRIAMMLFKS----------------------------------
MUL_0500|M.ulcerans_Agy99 DPMDTVLRIAMMLFKASRSGCRRFGEGCRRDEGRHHQHHDNPENHRVGVG
MAV_0728|M.avium_104 DPMDTVRRIATMLFAG----------------------------------
MAB_0499|M.abscessus_ATCC_1997 DPLRIASQIAYVLFPEAG--------------------------------
MLBr_00717|M.leprae_Br4923 TDADRLADSCAHALLDAIARVPEISKELADAMAIARQPPQ----------
Mb0672c|M.bovis_AF2122/97 -----------------
Rv0653c|M.tuberculosis_H37Rv -----------------
TH_4347|M.thermoresistible__bu -----------------
Mvan_5135|M.vanbaalenii_PYR-1 -----------------
Mflv_1618|M.gilvum_PYR-GCK -----------------
MSMEG_5838|M.smegmatis_MC2_155 -----------------
MMAR_4906|M.marinum_M -----------------
MUL_0500|M.ulcerans_Agy99 EIMHHVVGEVGGEGLGR
MAV_0728|M.avium_104 -----------------
MAB_0499|M.abscessus_ATCC_1997 -----------------
MLBr_00717|M.leprae_Br4923 -----------------