For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTPAQLRAYSAVVRLGSVRAAAAELGLSDAGVSMHVAALRKELDDPLFTRTGAGLAFTPGGLRLASRAVE ILGLQQQTAIEVTEAAHGRRLLRIAASSAFAEHAAPGLIELFSSRADDLSVELSVHPTSRFRELICSRAV DIAIGPASESSIGSDGSIFLRPFLKYQIITVVAPNSPLAAGIPMPALLRHQQWMLGPSAGSVDGEIATML RGLAIPESQQRIFQSDAAALEEVMRVGGATLAIGFAVAKDLAAGRLVHVTGPGLDRAGEWCVATLAPSAR QPAVSELVGFISTPRCIQAMIRGSGVGVTRFRPKVHVTLWS
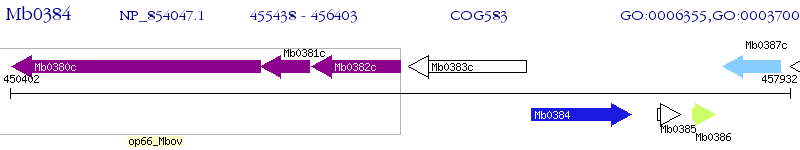
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0384 | - | - | 100% (321) | LysR family transcriptional regulator |
| M. bovis AF2122 / 97 | Mb2303c | - | 7e-10 | 26.09% (276) | LysR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1833 | - | 1e-08 | 25.81% (186) | LysR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0377 | - | 1e-177 | 99.69% (321) | LysR family transcriptional regulator |
| M. leprae Br4923 | MLBr_02041 | oxyR | 1e-05 | 25.41% (185) | putative LysR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3458c | - | 1e-09 | 24.90% (257) | LysR family transcriptional regulator |
| M. marinum M | MMAR_0653 | - | 1e-166 | 93.15% (321) | LysR family transcriptional regulator |
| M. avium 104 | MAV_2151 | - | 5e-10 | 25.71% (280) | LysR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_0742 | - | 1e-134 | 78.19% (321) | transcriptional regulatory protein |
| M. thermoresistible (build 8) | TH_0172 | - | 1e-129 | 73.52% (321) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY |
| M. ulcerans Agy99 | MUL_0119 | - | 1e-159 | 92.26% (310) | LysR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_3401 | - | 6e-07 | 25.00% (144) | LysR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0384|M.bovis_AF2122/97 --MTP-----AQLRAYSAVVRLGSVRAAAAELGLSDAGVSMHVAALRKEL
Rv0377|M.tuberculosis_H37Rv --MTP-----AQLRAYSAVVRLGSVRAAAAELGLSDAGVSMHVAALRKEL
MMAR_0653|M.marinum_M --MTP-----AQLRAYSAVVRLGSVRAAATELGLSDAGVSMHVAALRKEL
MUL_0119|M.ulcerans_Agy99 ------------------MVRLGSVRAAATELGLSDAGVSMHVAALRKEL
MSMEG_0742|M.smegmatis_MC2_155 --MTP-----AQLRAFSAVVRLGSVRAAAEELGVSDAGVSMHVAQLRKEL
TH_0172|M.thermoresistible__bu --MTP-----AQLRAFSAVVRLGSVRAAAQELGMSDAGISMHIAQLRKEL
MAB_3458c|M.abscessus_ATCC_199 MALPPGTPDLGVLDLLVSVAETGSLGAAARQHGISQPAASMRIRALERRL
MAV_2151|M.avium_104 MPLSPRMPELASFEAFLAIAETGSLGRAARELGLTQQAISRRLATMEAQI
Mflv_1833|M.gilvum_PYR-GCK --MDT-----HRLKYFLRIAEEGSITRAASLLGMAQPALSRQLQLLEEDL
MLBr_02041|M.leprae_Br4923 MSDKSYQPTIAGLRAFVAVVEKGHFSAAASFLGVRQSTLSQALAALESGL
Mvan_3401|M.vanbaalenii_PYR-1 --MDT-----RRLELLLALSRLGSMRAVADAHHLTTSTVSQQIAALARDT
: . * . .* : * : :
Mb0384|M.bovis_AF2122/97 DDPLFTRTGAGLAFTPGGLRLASRAVEILGLQQQTAIEVTEAAHG-RRLL
Rv0377|M.tuberculosis_H37Rv DDPLFTRTGAGLAFTPGGLRLASRAVEILGLQQQTAIEVTEAAHG-RRLL
MMAR_0653|M.marinum_M DDPLFTRTGAGLAFTPGGLRLASRAVEILGLQQQTAIEVTEAAHG-RRLL
MUL_0119|M.ulcerans_Agy99 DDPLFTRTGAGLAFTPGGLRLASGAVEILGLQQQTAIEVTEAAHG-RRLL
MSMEG_0742|M.smegmatis_MC2_155 DDPLFTRTASGLAFTPGGLRLASRAVEILGLQQQTAIEVTEAAHG-RRLL
TH_0172|M.thermoresistible__bu DDPLFHRTKNGLSFTPGGLRLASRAVDILGLQRQTAIEVTEAAHG-RRLL
MAB_3458c|M.abscessus_ATCC_199 RLVLLERGPTGSRLTDAGLAVVGYASPVLAAAHEFVTGVAALHSDQAPRL
MAV_2151|M.avium_104 GVTLAVRTTRGSQLTPAGLIVADWAARLLEVAHEIDAGLGSLRKEGRERI
Mflv_1833|M.gilvum_PYR-GCK GVALFRRNRRGVELTDAGERLRSSTAAPLRQLELAVQYAAFPLARLKRAM
MLBr_02041|M.leprae_Br4923 GVQLIERSTRRVFVTTQGRHLLPRAQAAIEAMDAFTAAAAGESDPLRGGM
Mvan_3401|M.vanbaalenii_PYR-1 GAALIEPEGRRVRLTPAGKRLADHAVTILAAVDRARLDLDPDAEP-AGTV
* .* * : : : :
Mb0384|M.bovis_AF2122/97 RIAASSAFAEHAAPGLIELFSSR-----ADDLSVELSVHPTSRFRELICS
Rv0377|M.tuberculosis_H37Rv RIAASSAFAEHAAPGLIELFSSR-----ADDLSVELSVHPTSRFRELICS
MMAR_0653|M.marinum_M RIAASSTFAEHAAPGLIELFSSR-----ADDLSVELSVHPTSQFRDLICS
MUL_0119|M.ulcerans_Agy99 RIAASSTFAEHAAPGLIELFSSR-----ADDLSVELSVHPTSQFRDLICA
MSMEG_0742|M.smegmatis_MC2_155 RIAASSAFAEHAAPGLIELFSSR-----ADDLSVELSVHPARQFGHLITS
TH_0172|M.thermoresistible__bu RIAASHAFAEHAAPGLIDLFSSR-----ADDLSVELTVEPVSRFRDLLES
MAB_3458c|M.abscessus_ATCC_199 VVAASRTIADHRIPLWLTALRAR-----HADVAVSLEVGNTQQVCELVRA
MAV_2151|M.avium_104 RVAASQTISEQLMPHWLLSLQAEATRRGEAAPQVILTATNSEHAIASVRD
Mflv_1833|M.gilvum_PYR-GCK LIGIPESAIDVLAAPLMTRLGSA-----FPDAQFAVTVGSTDHLVEAMLK
MLBr_02041|M.leprae_Br4923 RLGLIPTVAPYVLPTMLTGLTHR-----LPTLTLRVVEDQTEHLLTALRE
Mvan_3401|M.vanbaalenii_PYR-1 RVGGFATGIRVSLLPIVADLAVR-----YPEVDVVISEYEPVEAFKLLTA
:. : : : . : . :
Mb0384|M.bovis_AF2122/97 RAVDIAIG-PASESSIGSDGSIFLRPFLKYQIITVVAPNSPLAAG--IPM
Rv0377|M.tuberculosis_H37Rv RAVDIAIG-PASESSIGSDGSIFLRPFLKYQIITVVAPNSPLAAG--IPM
MMAR_0653|M.marinum_M RAVDIAIG-PASESSIGSDTSLFVRPFLKYQIITVVAPNSPLAVG--IPT
MUL_0119|M.ulcerans_Agy99 RAVDIAIG-PASESSIGSDTSLFVRPFLKYQIITVVAPNSPLAVG--IPT
MSMEG_0742|M.smegmatis_MC2_155 RAVDIALG-PAAP----ADPNLVVRAFLKYQIITVTTPDNALLER--GVT
TH_0172|M.thermoresistible__bu RAVDVTLG-PAVRE---PGPAIVVRPFLNYQIIAVTSPSNPLAAA--TPT
MAB_3458c|M.abscessus_ATCC_199 GGASLGFI-EGPHAPTGLGGEVLG----ADELVIVVGPGHKWAHRRKPVT
MAV_2151|M.avium_104 GSADLGFV-ENPGTPKGLGSCVVG----EDELVIVVPPTHKWARRSRVVT
Mflv_1833|M.gilvum_PYR-GCK GSVDIALI-NPVP-----DDRVFYREILTEEFVLVGGPDSPLEPE-KPLQ
MLBr_02041|M.leprae_Br4923 GALDAAMI-ALPVET----AGIAEIPIYDEDFVLALPPGHPLSGK-RRVP
Mvan_3401|M.vanbaalenii_PYR-1 DDLDLALTYDYNLAPASPGADLDAVPLWSIGWGLGVPSGTAADRG-----
. : : .
Mb0384|M.bovis_AF2122/97 PALLRHQQWMLGPSAGSVDG----EIATMLRGLAIPESQQRIFQSDAAAL
Rv0377|M.tuberculosis_H37Rv PALLRHQQWMLGPSAGSVDG----EIATMLRGLAIPESQQRIFQSDAAAL
MMAR_0653|M.marinum_M PSLLRQQQWMLGPPAGSVAG----EIATMLRDLAIPESQQRIFQSDAAAL
MUL_0119|M.ulcerans_Agy99 PSLLRQQQWMLGPPAGSVAG----EIATMLRDLAIPESQQRIFQSDAAAL
MSMEG_0742|M.smegmatis_MC2_155 PASLREQPWQLGPSGGNADG----EIAAMLRALAIPEARQRIFQSDAAAL
TH_0172|M.thermoresistible__bu IAQLRAQSWMLGPSAGGIDG----DVAAMLRRLEIPEAAQRIFQSDSAAQ
MAB_3458c|M.abscessus_ATCC_199 LRELAITPLLMREPGSGTRD----TVWEVLSEVCQPATPAAELGSAAAIK
MAV_2151|M.avium_104 ARELAQTPLVAREPHSGIRDSLTVALRKVLGEDMQQAAPVLELTSAAAMR
Mflv_1833|M.gilvum_PYR-GCK FADLVELPLVVPRSTTGLGQ----ALYNAALRAKVTIDYRITTDSTRVMV
MLBr_02041|M.leprae_Br4923 TTALAQLPLLLLDEGHCLRD---QTLDICRKSGVQAELANTRAASLATAV
Mvan_3401|M.vanbaalenii_PYR-1 LMTFADSTWIVNSRNTADET----AVRTLASLAGFTPTIAHQIDSLDLVE
: : *
Mb0384|M.bovis_AF2122/97 EEVMRVGGATLAIGFAVAKDLAAGRLVHVTGPGLDRAGEWCVATLAPSAR
Rv0377|M.tuberculosis_H37Rv EEVMRVGGATLAIGFAVAKDLAAGRLVHVTGPGLDRAGEWCVATLAPSAR
MMAR_0653|M.marinum_M EEVMRVGGATLTIGFAVAKDLAAGRLVHVTSPGLDKAGEWCVATLPPAAR
MUL_0119|M.ulcerans_Agy99 EEVMRVGGATLTIGFAVAKDLAAGRLVHVTSPGLDKAGEWCVATLPPAAR
MSMEG_0742|M.smegmatis_MC2_155 EEVRRVGGVTPTIGFAVAKDLAGGRLAQVKGPGLQVGGEWFATTLPASAR
TH_0172|M.thermoresistible__bu EEVQRIGGVTLSIGFAVAKDLAAGRLARIKG--VQATAEWCVSTLSSAAR
MAB_3458c|M.abscessus_ATCC_199 AAAATGLAPAVLSRLIAGPELSAGTLVEVKPADGTVFTRQFRAVWRPG-A
MAV_2151|M.avium_104 AAVLAGAGPAAMSRLAVADDLAVGRLHAVEIPK-LDLRRKFRAIWIGGRT
Mflv_1833|M.gilvum_PYR-GCK CLIESGLAYGVLPLSACGREFAESRLRYSRICEPVLTQQLGVAATEQLDL
MLBr_02041|M.leprae_Br4923 QCVTGGLGVTLLPQSAAPVESVRSKLGLAQFAAPCPGRRIGLAFRSASGR
Mvan_3401|M.vanbaalenii_PYR-1 DLIVAGFGVGLLP---LGRPTSPGVTVLPLQNPDVTLTAYAVTRRGRSAW
. . :
Mb0384|M.bovis_AF2122/97 QPAVSELVGFISTPRCIQAMIRGSGVGVTRFRPKVHVTLWS
Rv0377|M.tuberculosis_H37Rv QPAVSELVGFISTPRCIQAMIPGSGVGVTRFRPKVHVTLWS
MMAR_0653|M.marinum_M QPAVSELVRFITTPRCIQAMIRGSGVGVTRFRPKVHVTLWS
MUL_0119|M.ulcerans_Agy99 QPAVSELVRFITTPRCIQAMIRGSGVGVTRFRPKMHVTLWS
MSMEG_0742|M.smegmatis_MC2_155 QPAVSELLRFITTPRCTQAMIRGSGVGVSRFRPKVHVTLWS
TH_0172|M.thermoresistible__bu QPAASELVRFITTPRCTQAMIRGTSEGVTRFRPKVHVTLWS
MAB_3458c|M.abscessus_ATCC_199 PPTG-----------AARALVD--IAALVD-----------
MAV_2151|M.avium_104 PPAG-----------AIRDLLSHIIARTAASK---------
Mflv_1833|M.gilvum_PYR-GCK PRELVVKVG-NALREEIAALIRSGSWRADLLSPEPWSPRRV
MLBr_02041|M.leprae_Br4923 SASYR----------QIAKIIGELISTEHHVRLVA------
Mvan_3401|M.vanbaalenii_PYR-1 SPLRAILDR----MRPPAGLLQHPTWPRPEAAVANQSA---
::