For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNVGVHHRRCAIRHVTTFAAALVSVSGIAEIWGRPVSVGFQPLTSSEHRLDSNPGEERGRTQRTGRGDRM VTDYNQGEIAEQYKKTKAIPVRTRIEAYSFLKHIGAVTGEKVVDIACGAGDYTRVMRRAGAARVVGFDIS EKMIGLAREQEAHEPLGIEYFVADASQEVAQQDFDLAISAYLLVYARDRDELARICRGVACRVRPGGRFV TLTTNPGLYTFGRVPDYRKYGFQIKLADAAFEGAPIELTAFVDGAPLVIENYYLPIAAYEAALRQAGFHD FVVHVPELAAAPQGEDEGDFWDDYLNYPPAIVIECVRD
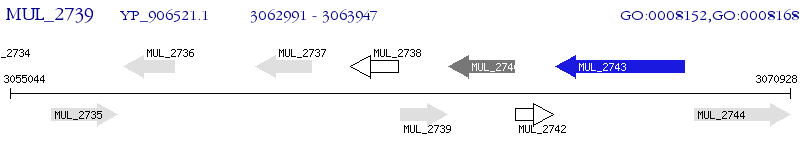
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_2739 | - | - | 100% (318) | methylase |
| M. ulcerans Agy99 | MUL_1801 | - | 3e-06 | 29.59% (98) | methyltransferase |
| M. ulcerans Agy99 | MUL_1216 | tam | 6e-06 | 31.25% (96) | trans-aconitate methyltransferase Tam |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1440c | - | 6e-07 | 30.56% (108) | putative methyltransferase |
| M. gilvum PYR-GCK | Mflv_2768 | - | 6e-06 | 32.08% (106) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv1405c | - | 6e-07 | 30.56% (108) | putative methyltransferase |
| M. leprae Br4923 | MLBr_00551 | - | 2e-05 | 28.57% (98) | putative methyltransferase |
| M. abscessus ATCC 19977 | MAB_4018 | - | 4e-05 | 28.95% (152) | similarity with UbiE/COQ5 methyltransferase |
| M. marinum M | MMAR_3525 | - | 1e-141 | 97.99% (249) | methylase |
| M. avium 104 | MAV_3373 | - | 3e-08 | 32.80% (125) | methyltransferase, UbiE/COQ5 family protein |
| M. smegmatis MC2 155 | MSMEG_3286 | - | 6e-08 | 28.14% (199) | methyltransferase |
| M. thermoresistible (build 8) | TH_1636 | - | 2e-06 | 27.54% (167) | POSSIBLE METHYLTRANSFERASE (METHYLASE) |
| M. vanbaalenii PYR-1 | Mvan_4690 | - | 1e-06 | 31.97% (122) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_2739|M.ulcerans_Agy99 -----------MNVGVHHRRCAIRHVTTFAAALVSVSGIAEIWGRPVSVG
MMAR_3525|M.marinum_M --------------------------------------------------
Mb1440c|M.bovis_AF2122/97 --------------------------------------------------
Rv1405c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00551|M.leprae_Br4923 --------------------------------------------------
MAV_3373|M.avium_104 --------------------------------------------------
MSMEG_3286|M.smegmatis_MC2_155 --------------------------------------------------
TH_1636|M.thermoresistible__bu ---------------------------------------------VRESR
MAB_4018|M.abscessus_ATCC_1997 -------------------------------MLHHPTLADSLIDPARIDH
Mvan_4690|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_2768|M.gilvum_PYR-GCK MTTLAENSAPSSETTEEFAQRIVGALDAASLALLVSIGHQTHLFDTMANL
MUL_2739|M.ulcerans_Agy99 FQPLTSSEHRLDSNPGEERGRTQRTGRGDRMVTDYNQGEIAEQYKKTKAI
MMAR_3525|M.marinum_M ------------------------------MVTDYNQGEIAERYKKTKAI
Mb1440c|M.bovis_AF2122/97 -------------------MTIDTPAREDQTLAATHRAMWALGDYALMAE
Rv1405c|M.tuberculosis_H37Rv -------------------MTIDTPAREDQTLAATHRAMWALGDYALMAE
MLBr_00551|M.leprae_Br4923 -------------------MNLYTPIHEDQSLAAIHRAMWALGDYALMAE
MAV_3373|M.avium_104 -------------------MTLDTTT-ADASVAARHRTVWALGDYALMAE
MSMEG_3286|M.smegmatis_MC2_155 --------------------------MPDTVGPQPQYETFADEFLDHARD
TH_1636|M.thermoresistible__bu DQVFRKLNEKLTQHGQKFIYSYASRRLDDDDVVFLNYGYEEDPPMGLPLS
MAB_4018|M.abscessus_ATCC_1997 PPRRDHGYLDVLGPESDEPVSVANRFMQSPLLAAIYERAWRPMFTRGFSY
Mvan_4690|M.vanbaalenii_PYR-1 ---------------------MVDRANVAQVQAWNGDSGRAWVTLQHVLD
Mflv_2768|M.gilvum_PYR-GCK PPSTSTQIADAAGLNERYVREWLGGVAAGRIVDYDPAAQTFSLPAHRAAV
MUL_2739|M.ulcerans_Agy99 PVR----TRIEAYSFLKHIGAVTGEKVVDIACGAGD---------YTRVM
MMAR_3525|M.marinum_M PVR----TRIEAYSFLKHIGDVTGEKVVDIACGAGD---------YARVM
Mb1440c|M.bovis_AF2122/97 EVM----APLGP-ILVAAAGIGPGVRVLDVAAGSGN---------ISLPA
Rv1405c|M.tuberculosis_H37Rv EVM----APLGP-ILVAAAGIGPGVRVLDVAAGSGN---------ISLPA
MLBr_00551|M.leprae_Br4923 EVM----APLGP-ILVTATGIRPGMQVLDVAAGSGN---------ISLPA
MAV_3373|M.avium_104 EVM----APLGP-TLVEAAGIGPGVRVLDVAAGSGN---------ITLPA
MSMEG_3286|M.smegmatis_MC2_155 GFYN---AHYDRPACLALIGDVAGRTILDAACGPGL---------YAEEL
TH_1636|M.thermoresistible__bu ETDEPNRYSIQLYHRTAGQADLTGKRVLEVGCGHGGG--------ASYLM
MAB_4018|M.abscessus_ATCC_1997 GGKS---TLKAHTALMDEIAGRGDHKILDVACGPGLY--------TRELA
Mvan_4690|M.vanbaalenii_PYR-1 RTLK---PFEDLIVDSVAAAAGPREQILDLGCGTGAT--------TLALA
Mflv_2768|M.gilvum_PYR-GCK LTRTAGPDNLARVTQFIPLLSEVEQKVIACFRDGGGLSYAEYPRFHTLMA
:: . *
MUL_2739|M.ulcerans_Agy99 RRAGAAR-VVGFDISEKMIGLAREQEAHEPLGIEYFVADAS-QEVAQQDF
MMAR_3525|M.marinum_M RRAGAAR-VVGFDISEKMIGLAREQEAHEPLGIEYFVADAS-QEVAQQDY
Mb1440c|M.bovis_AF2122/97 AKTGAT--VISTDLTPELLQRSQARAAQQGLTLQYQEANAQALPFADDEF
Rv1405c|M.tuberculosis_H37Rv AKTGAT--VISTDLTPELLQRSQARAAQQGLTLQYQEANAQALPFADDEF
MLBr_00551|M.leprae_Br4923 AKTGAT--IVSTDLTPELLQRSQARATELGLTLEYREADAQALPFGDGEF
MAV_3373|M.avium_104 AAAGAS--VVSTDLTPELLQRSRDRAAGMGLTIDYREANAQALPFGDGEF
MSMEG_3286|M.smegmatis_MC2_155 AKRGAS--LIGFDQSPRMVELARARVPSGDFRVHDLAGPLDWLPSQS--V
TH_1636|M.thermoresistible__bu RTLGPMS-YVGLDLNPAGIEFCRKKHRLP--GLEFVVGDAQDLPFGAESF
MAB_4018|M.abscessus_ATCC_1997 AQLGTAGVCIGLDLSGPMLR--RAVRDNSAERVDYIRGSAHSLPFADATF
Mvan_4690|M.vanbaalenii_PYR-1 ERLGAHGGCTGIDISEPMIHAARTRARDRNLPVEFVVGDAQSFPFQPNTF
Mflv_2768|M.gilvum_PYR-GCK EQSGEVFDAALVDGILPMAEGLTDRLRQGVDVADFGCGSGHAVNVMAKAF
* * . .
MUL_2739|M.ulcerans_Agy99 DLAISAYLLVYARDRDELARICRGVACRVRPGGRFVTLTTNPGLYTFGRV
MMAR_3525|M.marinum_M DLAISAYLLVYARDRDELARMCRGVACRVRPGGRFVTLTTNPGLYTFGRV
Mb1440c|M.bovis_AF2122/97 DTVISAIGVMFAPDHQ---AAADELVRVCRPGGTIGVISWTCEGFFGRML
Rv1405c|M.tuberculosis_H37Rv DTVISAIGVMFAPDHQ---AAADELVRVCRPGGTIGVISWTCEGFFGRML
MLBr_00551|M.leprae_Br4923 DVVMSAIGVMFAPDHR---RAASELVRVCRPNGTIGVISWTQEGFFGRML
MAV_3373|M.avium_104 DAVVSAIGVQFAPDHQ---RAADELVRVCRPGGRIGLISWTPEGFFGRML
MSMEG_3286|M.smegmatis_MC2_155 DAVLLALALEYVDDRT---AMLREFHRVLRPAGALVLSRMHPTGDWLRHG
TH_1636|M.thermoresistible__bu DAVINIESSHLYPRFS---RFLSEVARVLRPGGHFLYADARQPHEFAEWE
MAB_4018|M.abscessus_ATCC_1997 DTVVCLAALYLIPDPE---QAVRELCRVAGPDGQIVVFTSLRTRAAS---
Mvan_4690|M.vanbaalenii_PYR-1 DVMVSRFGVMFFDDPT---TAFRNLRRATRPGGRLTFVCWRDPTDNPFMS
Mflv_2768|M.gilvum_PYR-GCK PASR-FTGIDFSEEGLAAGRDEARRLGLTNASFNATDVATYDAAESFDVI
.
MUL_2739|M.ulcerans_Agy99 PDYRKYG--FQIKLADAAFEGAPI-ELTAFVDG--APLVI-ENYYLPIAA
MMAR_3525|M.marinum_M PDYRKYG--FQIKLADAAFEGAPI-ELTAFVDG--APLVI-ENYYLPIAA
Mb1440c|M.bovis_AF2122/97 ATIRPYRPSVSADLPPSALWGREA-YVTGLLGDGVTGLKT-ARGLLEVKR
Rv1405c|M.tuberculosis_H37Rv ATIRPYRPSVSADLPPSALWGREA-YVTGLLGDGVTGLKT-ARGLLEVKR
MLBr_00551|M.leprae_Br4923 ATIRPYRPSLSAALPPAALWGREA-YIRGLLDDTVTNVKT-ERRQLEVKR
MAV_3373|M.avium_104 ATIRPYRPSLSHPVPPAALWGRPG-YVRALLGERIGEITT-ARGMLPVNR
MSMEG_3286|M.smegmatis_MC2_155 GNYFETR-----VVEETWSRGWNVRYWLAPLERTCSELRD-AGFLIEELR
TH_1636|M.thermoresistible__bu AALAAAPLDMVSARVINAEVVRGMEQILPVWEDVIERVAP-AFLRRRVRE
MAB_4018|M.abscessus_ATCC_1997 --------------LPGVTTAMRIGGFRAFGRDEVTGWLR-AQG------
Mvan_4690|M.vanbaalenii_PYR-1 TAERAAAQLLSDLKPGIAGSPGPFAFADAHRTHSILRAGGWTAVDIRPVD
Mflv_2768|M.gilvum_PYR-GCK TAFDAIHDQAHPARVLANIHAALRPGGTFLMVDIKSSSRLENNIGVPFAP
MUL_2739|M.ulcerans_Agy99 YEAALRQAGFH---------------DFVVHVPELAAAPQGEDEGDFWDD
MMAR_3525|M.marinum_M YEAALRQAGFH---------------DFVVHVPELAAAPQGEDEGDFWDD
Mb1440c|M.bovis_AF2122/97 FDTAQAVHDYFKNNYGPTIEAYAHIGDNAVLAAELDRQLVELAAQYLSDG
Rv1405c|M.tuberculosis_H37Rv FDTAQAVHDYFKNNYGPTIEAYAHIGDNAVLAAELDRQLVELAAQYLSDG
MLBr_00551|M.leprae_Br4923 FNSAEGVHNYFKNHYGPTIEAYTNIGDNSVLAAELDAQLTELAQKYLTNS
MAV_3373|M.avium_104 FGSAEDVHAYFKQHYGPTIEAYANIGHNRVLAAELDAQLVELAAQHLSDG
MSMEG_3286|M.smegmatis_MC2_155 EPRPTEEAARR--------------------DPQAYQRLAQAPEGFMTIR
TH_1636|M.thermoresistible__bu FAP----------------------------ARRAWEELRTEGSTEYRMY
MAB_4018|M.abscessus_ATCC_1997 -------------------------------WTDIDQTVTGQGQFIRARR
Mvan_4690|M.vanbaalenii_PYR-1 VTCTMQESDLLPYLTQMGPVGRTLRAVDARTRAQVITSLREAYTEFIVDG
Mflv_2768|M.gilvum_PYR-GCK YLYTVSAMHCMSVSLGLDGDGLG----TCWGREVATTMLADAGFGDVQVR
MUL_2739|M.ulcerans_Agy99 YLNYPPAIVIECVRD-
MMAR_3525|M.marinum_M YLNYPPAIVIECVRD-
Mb1440c|M.bovis_AF2122/97 VMEWEYLLLTAEKR--
Rv1405c|M.tuberculosis_H37Rv VMEWEYLLLTAEKR--
MLBr_00551|M.leprae_Br4923 TMGWEYLLLTAQKH--
MAV_3373|M.avium_104 TMGWEYLLVTAEKRSG
MSMEG_3286|M.smegmatis_MC2_155 AVPDFRLRRPAAGR--
TH_1636|M.thermoresistible__bu CFTKPPEHADIP----
MAB_4018|M.abscessus_ATCC_1997 AVALRVAT--------
Mvan_4690|M.vanbaalenii_PYR-1 EVRYPAACWMVDARG-
Mflv_2768|M.gilvum_PYR-GCK EIESDPINYYYIARK-
.