For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSLLDTLGTVRSVAEALHLSPSTVSQQLALLEGETRTRLLERAGRRVRLTPTGLLLARRGREIIDRMAAV EAELRELSDEPVGTVRLGLFQSAIPTLGIPASARLADTHPHLHLELIELEPHESGAALRASEVDVIVTTT DYMEFPWGDDLDIVAIGTDPIVLVLPQDHRLGRRPVVDLATCSDETWVCDRPDSYMAELTLRLCRQSGFE PRVAGRFSTVSLLVHHVQTQGSVALLPALAVGPEQKVLTRELTLPVHRNVAAAFRRSAARPAVHAVVDAL RDHPEIPALTER*
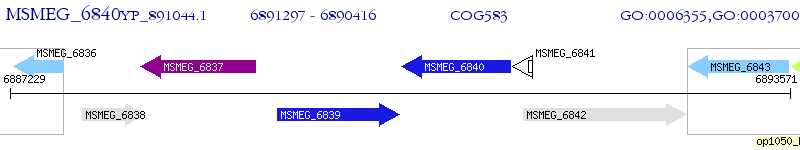
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6840 | - | - | 100% (293) | LysR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_3260 | - | 4e-27 | 32.40% (287) | LysR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_1909 | - | 1e-19 | 29.01% (293) | putative HTH-type transcriptional regulator YnfL |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0121 | oxyS | 4e-08 | 28.21% (156) | oxidative stress response regulatory protein OXYS |
| M. gilvum PYR-GCK | Mflv_1356 | - | 5e-18 | 28.07% (285) | LysR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0117 | oxyS | 4e-08 | 28.21% (156) | oxidative stress response regulatory protein OXYS |
| M. leprae Br4923 | MLBr_02041 | oxyR | 1e-14 | 29.71% (276) | putative LysR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_2680c | - | 8e-20 | 29.08% (282) | LysR family transcriptional regulator |
| M. marinum M | MMAR_4117 | - | 3e-26 | 35.81% (296) | putative regulatory protein |
| M. avium 104 | MAV_1538 | - | 3e-14 | 25.64% (234) | transcriptional regulator, LysR family protein |
| M. thermoresistible (build 8) | TH_3083 | - | 3e-05 | 27.04% (196) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_3983 | - | 4e-26 | 35.47% (296) | putative regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_3401 | - | 3e-25 | 32.78% (302) | LysR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4117|M.marinum_M -------------------------MLNLQRVVVLHELQRRGTLAAVAKA
MUL_3983|M.ulcerans_Agy99 -------------------------MLNLQRVVVLHELQRRGTLAAVAKA
Mb0121|M.bovis_AF2122/97 --------------------------MLFRQLEYFVAVAQERHFARAAEK
Rv0117|M.tuberculosis_H37Rv --------------------------MLFRQLEYFVAVAQERHFARAAEK
MSMEG_6840|M.smegmatis_MC2_155 ---------------------------------MLSLLDTLGTVRSVAEA
Mvan_3401|M.vanbaalenii_PYR-1 --------------------------MDTRRLELLLALSRLGSMRAVADA
Mflv_1356|M.gilvum_PYR-GCK MVSAVTGRPTNDPPHTWRHRTIKEPPMELRHLRYFLAVAETCHFGQAAEQ
MAV_1538|M.avium_104 --------------------------MAVGDYEWFITLAELQHVTAAAEQ
TH_3083|M.thermoresistible__bu --------------------------------------------------
MAB_2680c|M.abscessus_ATCC_199 -------------------MDQWTSQLAPQLRALVELAAHDGHMTQAALT
MLBr_02041|M.leprae_Br4923 -------------------MSDKSYQPTIAGLRAFVAVVEKGHFSAAASF
MMAR_4117|M.marinum_M LSYSPSAVSQQLAQLERECGMALIEPVGRGVRLTDAGEALVEHTRLAIAT
MUL_3983|M.ulcerans_Agy99 LSYSPSAVSQQLAQLERECGMALIEPVGRGVRLTDAGEALVEHTRLAIAT
Mb0121|M.bovis_AF2122/97 CYVSQPALSSAIAKLERELNVTLINRGHSFEGLTREGERLVVWAKRILAE
Rv0117|M.tuberculosis_H37Rv CYVSQPALSSAIAKLERELNVTLINRGHSFEGLTREGERLVVWAKRILAE
MSMEG_6840|M.smegmatis_MC2_155 LHLSPSTVSQQLALLEGETRTRLLERAGRRVRLTPTGLLLARRGREIIDR
Mvan_3401|M.vanbaalenii_PYR-1 HHLTTSTVSQQIAALARDTGAALIEPEGRRVRLTPAGKRLADHAVTILAA
Mflv_1356|M.gilvum_PYR-GCK LHIAQPALSYAIRQLENSLDATLFTRTTRHVALTPAGEFLRVEATRILNG
MAV_1538|M.avium_104 LHVAQPTLTRMLARLEQRLGVALFDRHGRRLSLNTYGRIFYEHARRAQLE
TH_3083|M.thermoresistible__bu --------------------------------------------------
MAB_2680c|M.abscessus_ATCC_199 LGIPQSSMSRRIHALQAALGVPLLIHDGRTVRLTPEARLLAARTREPLAE
MLBr_02041|M.leprae_Br4923 LGVRQSTLSQALAALESGLGVQLIERSTRRVFVTTQGRHLLPRAQAAIEA
MMAR_4117|M.marinum_M LERAEALLAASQGRVAGRLRVAAFQTLLLSLLPRVLDALARDYPDLRVEV
MUL_3983|M.ulcerans_Agy99 LERAEALLAASQGRVAGRLRVAAFRTLLLSLPPRVLDALARDYPDLRVEV
Mb0121|M.bovis_AF2122/97 HAAFKAEVDAVRSGITGTLRLGTVPTASTTAS-LVLSAFCSAHPLAKVQV
Rv0117|M.tuberculosis_H37Rv HAAFKAEVDAVRSGITGTLRLGTVPTASTTAS-LVLSAFCSAHPLAKVQV
MSMEG_6840|M.smegmatis_MC2_155 MAAVEAELRELSDEPVGTVRLGLFQSAIPTLGIPASARLADTHPHLHLEL
Mvan_3401|M.vanbaalenii_PYR-1 VDRARLDLDP-DAEPAGTVRVGGFATGIRVSLLPIVADLAVRYPEVDVVI
Mflv_1356|M.gilvum_PYR-GCK VDAAQRGVRRIAAGRSGLLRLGLTGTAAFSHLPKIARIVRQELPAVALDI
MAV_1538|M.avium_104 LDSARRAIADLADPAVGEIRLAYLSSFGSTVVPRLIARFKESSPRVTFTL
TH_3083|M.thermoresistible__bu ------------------VRITFPGMSTHPHVARLARVVRSQRPGIRLEL
MAB_2680c|M.abscessus_ATCC_199 LDHTLSEVTGDADPERGTVRFGFPLTMGSGHIPDLLAAFRHRHPGIRVLL
MLBr_02041|M.leprae_Br4923 MDAFTAAAAGESDPLRGGMRLGLIPTVAPYVLPTMLTGLTHRLPTLTLRV
:*. . * . :
MMAR_4117|M.marinum_M AQREADEATSGLLAGTFDVVLGEEYPGGPLPPSPSLDRVDLRTDELFLAM
MUL_3983|M.ulcerans_Agy99 AQREADEATSGLLAGTFDVVLGEEYPGGPLPPSPSLDRVDLRTDELFLAM
Mb0121|M.bovis_AF2122/97 CSRLAATELYRRLR---EFELDAVIVHPETQDSDDVDLVPLYEEQYVLLS
Rv0117|M.tuberculosis_H37Rv CSRLAATELYRRLR---EFELDAVIVHPETQDSDDVDLVPLYEEQYVLLS
MSMEG_6840|M.smegmatis_MC2_155 IELEPHESGAALRASEVDVIVTTTD-YMEFPWGDDLDIVAIGTDPIVLVL
Mvan_3401|M.vanbaalenii_PYR-1 SEYEPVEAFKLLTADDLDLALTYDYNLAPASPGADLDAVPLWSIGWGLGV
Mflv_1356|M.gilvum_PYR-GCK HADLLTPDQCEGLR----TGALDVGVLRPPAVGGDVATRTIAAEPLVLAV
MAV_1538|M.avium_104 EEGAAESIADRVRS-----GGVDIGVVSPRPVERTLAWRSLFRQRLGVAV
TH_3083|M.thermoresistible__bu SSQNFAQPAMQKLVR----SETDIALGRWDLIPADVSSRIVMRDELVVAL
MAB_2680c|M.abscessus_ATCC_199 KQAHGSELGAQLLTG-------DLDLAVVIPAPERLHHITIGAQHIHIAV
MLBr_02041|M.leprae_Br4923 VEDQTEHLLTALREG-----ALDAAMIALPVETAGIAEIPIYDEDFVLAL
: : :
MMAR_4117|M.marinum_M PR--IGPWSDATTLSDLEYASWALDP--EPTVPGRWARSVCRAAGFEPDV
MUL_3983|M.ulcerans_Agy99 PR--IGPWSDATTLSDLEYASWALDP--EPTVPGRWARSVCRAAGFEPDV
Mb0121|M.bovis_AF2122/97 PADMLPPGTSTLVWRDAAQLPLALLT--ADMRDRQVIDAAFADHAVSAIP
Rv0117|M.tuberculosis_H37Rv PADMLPPGTSTLVWRDAAQLPLALLT--ADMRDRQVIDAAFADHAVSAIP
MSMEG_6840|M.smegmatis_MC2_155 PQDHRLGRRPVVDLATCSDETWVCDR--PDSYMAELTLRLCRQSGFEPRV
Mvan_3401|M.vanbaalenii_PYR-1 PS----GTAADRGLMTFADSTWIVNS--RNTADETAVRTLASLAGFTPTI
Mflv_1356|M.gilvum_PYR-GCK SADHRLAAEPVVSLADLRSEPFIGYTG-QDSAVNDAVTRSCRAAGFVPHR
MAV_1538|M.avium_104 PDGHRFARGAAVSMVDLADEPFVTMH--PGFGMRRLLDELCAAAQFRPRV
TH_3083|M.thermoresistible__bu PEDHALAGRGRVSAGELAGESFVSLPPYEGSVLPDRLRRLAHANGFVPEV
MAB_2680c|M.abscessus_ATCC_199 PEDHRLAGAAQVRLRELSGETFIANP--PSYNLRQLTERWCREAGYAPNV
MLBr_02041|M.leprae_Br4923 PPGHPLSGKRRVPTTALAQLPLLLLD--EGHCLRDQTLDICRKSGVQAEL
. . .
MMAR_4117|M.marinum_M R-FDGVDLLVHLQMVQAGLAVAILP-ALLGQDRMRG-----VRLAPLPAS
MUL_3983|M.ulcerans_Agy99 R-FDGVDLLVHLQMVQAGLAVAILP-ALLGQDRMRG-----VRLAPLPAS
Mb0121|M.bovis_AF2122/97 Q-VETDSVASLFAQVATGNWASIVPHTWLWAMPMSGPTGGEIRAVELVDP
Rv0117|M.tuberculosis_H37Rv Q-VETDSVASLFAQVATGNWASIVPHTWLWAMPMSGPTGGEIRAVELVDP
MSMEG_6840|M.smegmatis_MC2_155 A-GRFSTVSLLVHHVQTQGSVALLPALAVGPE-------QKVLTRELTLP
Mvan_3401|M.vanbaalenii_PYR-1 A-HQIDSLDLVEDLIVAGFGVGLLP---LGRPTSPG-----VTVLPLQNP
Mflv_1356|M.gilvum_PYR-GCK E-HSAPGTAVLLALVAADLGVSVVP-ASVRALPLHG-----VVFRDLVDG
MAV_1538|M.avium_104 A-LQAPNLTTAASLVAAGLGISLVP--IDGSSYPSG-----VSVKPLADA
TH_3083|M.thermoresistible__bu V-QIAPDTQTALALVSAQVGCHLTFASVARNATDPH-----VVFVPLEGA
MAB_2680c|M.abscessus_ATCC_199 T-IEVTEFSTIRELIGRGLGVALLP---HDDRALPG-----IREIPLAGD
MLBr_02041|M.leprae_Br4923 ANTRAASLATAVQCVTGGLGVTLLPQSAAPVESVRS----KLGLAQFAAP
: : : :
MMAR_4117|M.marinum_M --PKRQLFTL--TRTSRTAHPAVVAFRTALAEGFDAQHQYD---------
MUL_3983|M.ulcerans_Agy99 --PKRQLFTL--TRTSRTAHPAVVAFRTALAEGFDAQHQYD---------
Mb0121|M.bovis_AF2122/97 --VLKAQIAL--ATNALGPGSPVARALITCAQALALNEFFDTQLRGITRR
Rv0117|M.tuberculosis_H37Rv --VLKAQIAL--ATNALGPGSPVARALITCAQALALNEFFDTQLRGITRR
MSMEG_6840|M.smegmatis_MC2_155 --VHRNVAAA--FRRS-AARPAVHAVVDALRDHPEIPALTER--------
Mvan_3401|M.vanbaalenii_PYR-1 D-VTLTAYAV--TRRGRSAWSPLRAILDRMRPPAGLLQHPTWPRPEAAVA
Mflv_1356|M.gilvum_PYR-GCK ---GTVDLAL--AWRDGVDNPVVDAVVAVLAGALTSLDHPALQEQS----
MAV_1538|M.avium_104 --DAYRDVGM--IWDSGRPLPRSARDFIAGAAAVGHGGW-----------
TH_3083|M.thermoresistible__bu A-EADVDVDLRAAWRRDDRSPAVHAVLECLFDDAGVENR-----------
MAB_2680c|M.abscessus_ATCC_199 G-YHRAIALAWGTATRAAPTRRLNDFLLQHFSQAEVNSAAIRKG------
MLBr_02041|M.leprae_Br4923 CPGRRIGLAFRSASGRSASYRQIAKIIGELISTEHHVRLVA---------
MMAR_4117|M.marinum_M ----
MUL_3983|M.ulcerans_Agy99 ----
Mb0121|M.bovis_AF2122/97 R---
Rv0117|M.tuberculosis_H37Rv R---
MSMEG_6840|M.smegmatis_MC2_155 ----
Mvan_3401|M.vanbaalenii_PYR-1 NQSA
Mflv_1356|M.gilvum_PYR-GCK ----
MAV_1538|M.avium_104 ----
TH_3083|M.thermoresistible__bu ----
MAB_2680c|M.abscessus_ATCC_199 ----
MLBr_02041|M.leprae_Br4923 ----