For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAQALHVQIARDLRDRIRRGELGLGTALPSESQLCAQWKSSRGPVRQALTTLRAEGLITGGPGKPPVVCS TAVGQPFDTLLSYSAWAHSIGRTPGQHTLELSLRRADEHTAASLHVEVGSPVVQLLRLRLLDGSPAMLER AAFVERIGRTLFDFDCDSGSIWAFLQSRGVQLATASHTIDAVGADPVDATNLQVPEGTPLLRQRRTTRAA DGEVIEFHDDRYLPGLVTFTLENTLETRTTLARNATR
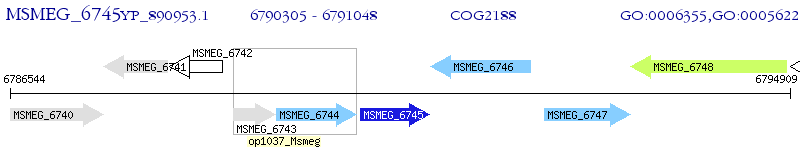
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6745 | - | - | 100% (247) | transcriptional regulator, GntR family protein |
| M. smegmatis MC2 155 | MSMEG_1227 | - | 6e-20 | 34.25% (219) | transcriptional regulator, GntR family protein |
| M. smegmatis MC2 155 | MSMEG_0286 | - | 9e-17 | 32.12% (193) | GntR-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0816c | - | 1e-09 | 29.91% (224) | GntR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2652 | - | 2e-16 | 28.51% (235) | regulatory protein GntR, HTH |
| M. tuberculosis H37Rv | Rv0792c | - | 2e-09 | 29.91% (224) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2239 | - | 2e-18 | 30.62% (209) | GntR family transcriptional regulator |
| M. marinum M | MMAR_4842 | - | 1e-11 | 26.43% (227) | regulatory protein |
| M. avium 104 | MAV_5032 | - | 2e-20 | 35.62% (219) | GntR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_0099 | - | 1e-10 | 27.03% (222) | transcriptional regulator, GntR family protein |
| M. ulcerans Agy99 | MUL_0525 | - | 1e-13 | 29.07% (227) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_3927 | - | 4e-19 | 31.20% (234) | UbiC transcription regulator-associated domain-containing |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0816c|M.bovis_AF2122/97 MTSVKLDLDAADLRISRGSVPASTQLAEALKAQIIQQRLPRGGRLPSERE
Rv0792c|M.tuberculosis_H37Rv MTSVKLDLDAADLRISRGSVPASTQLAEALKAQIIQQRLPRGGRLPSERE
Mflv_2652|M.gilvum_PYR-GCK -MSTTEADLRGYSAKMPAGTPLYMSIASDVRDLIASEALGPHTLLPSERE
Mvan_3927|M.vanbaalenii_PYR-1 ---------------MPAGTPLYMTIAADVRDRIATEQLGPHTLLPSERE
MAB_2239|M.abscessus_ATCC_1997 -------MAAAEPTPLRRPRADQARLVADVLRHQIHAGGYAEGGLPSEHA
MAV_5032|M.avium_104 ------------------------MLRSQILEGAYGGLAAARPALPSESA
MMAR_4842|M.marinum_M --MEGCRVLQLGQIDRTDDKPPYRQIASMLREAISSSRFPPGERLPSEAE
MUL_0525|M.ulcerans_Agy99 --MEGSQVLQLGQIDRADDKPPYRQIASMLRGAIISGELEAGGRLPSEAE
MSMEG_6745|M.smegmatis_MC2_155 -----------------MAQALHVQIARDLRDRIRRGELGLGTALPSESQ
TH_0099|M.thermoresistible__bu -------------MTVSPADHRYLQVARILRKEIVDGVYPVGSQLPTEHE
: : **:*
Mb0816c|M.bovis_AF2122/97 LIDRSGLSRVTVRAAVGMLQRQGWLVRRQGLGTFVADP-----------V
Rv0792c|M.tuberculosis_H37Rv LIDRSGLSRVTVRAAVGMLQRQGWLVRRQGLGTFVADP-----------V
Mflv_2652|M.gilvum_PYR-GCK LAEQHGVSRMTARQALSLLESEGVVYRKPPRGTFVAEP-----------R
Mvan_3927|M.vanbaalenii_PYR-1 LAEQHGVSRMTARQALSLLESEGVVYRKPPRGTFVAEP-----------R
MAB_2239|M.abscessus_ATCC_1997 LAAEFFVSRNTVREALTTLKDEGLIERGPRTGTHVALR-----------K
MAV_5032|M.avium_104 LAAELGVSRNAIREALELLRGEGLITRVQGSGTFVTGA-----------K
MMAR_4842|M.marinum_M LIEHFGVARMTVRQAMQELRSDGLVISQHGRGVFVRPTPPIRRLASDRFA
MUL_0525|M.ulcerans_Agy99 LIEYFGVARMTVRQAVQELRTEGLVISEHGRGVFVRPAPPIRWLASDRFA
MSMEG_6745|M.smegmatis_MC2_155 LCAQWKSSRGPVRQALTTLRAEGLITGGPGKPPVVCST-----------A
TH_0099|M.thermoresistible__bu LSERFSVSRYTIREALRRLREDNLISSRPRAGTMVVPRPS---------S
* :* . * *: *. :. : *
Mb0816c|M.bovis_AF2122/97 EQELSCGVRTITEVLLSCGVTPQVDVLSHQTGPAPQRISETLGLVE---V
Rv0792c|M.tuberculosis_H37Rv EQELSCGVRTITEVLLSCGVTPQVDVLSHQTGPAPQRISETLGLVE---V
Mflv_2652|M.gilvum_PYR-GCK VRFHIGSFS---EEVARMGRRPAAQLLWAEHQAATPAVRRALGLDEGAAV
Mvan_3927|M.vanbaalenii_PYR-1 VRFHVGSFS---EEVARMGRRPAARQLWAEHQLASPAVRRALGLAEGAAV
MAB_2239|M.abscessus_ATCC_1997 YEHGLDALLGLKETFKGYG-EVRNEVRVIQEVNAPPAVAHRLRLDPGARV
MAV_5032|M.avium_104 LRQDLNRLKGLAESLAGHRLPVENKVLSIRESTATPFVAAKLQLPENAPI
MMAR_4842|M.marinum_M RQHRAEGKAAFSVEAEKSGYSPQVDNIAVSREKPNSFIAEQLRISADDDI
MUL_0525|M.ulcerans_Agy99 RQHRANGKAAFTVEAERSGHTPQVDNITVSREKPDSVVAERLRLTADDEV
MSMEG_6745|M.smegmatis_MC2_155 VGQPFDTLLSYSAWAHSIGRTPGQHTLELSLRRADEHTAASLHVEVGSPV
TH_0099|M.thermoresistible__bu HAYAQDVMSINDLLAFATGAKFAIESVAMVTIDDELADRTGLTVDEQWLA
* :
Mb0816c|M.bovis_AF2122/97 LCIRRRIRTGDQPLALVTAYLPPGVGPAVEPLLSGSADTETTYAMWERRL
Rv0792c|M.tuberculosis_H37Rv LCIRRRIRTGDQPLALVTAYLPPGVGPAVEPLLSGSADTETTYAMWERRL
Mflv_2652|M.gilvum_PYR-GCK HVFHRLRSVDEVPFALETTFLPAELTPGI---LDMTED-GSLWAVLRSKY
Mvan_3927|M.vanbaalenii_PYR-1 HVFHRLRSVDDVPFALETTYLPADLTPGI---LEITDE-GSLWAVLRSRY
MAB_2239|M.abscessus_ATCC_1997 IFIERLRYLGDLPLSLDKTYLTPDVGEAV---IEHQLETNDVFALIERVT
MAV_5032|M.avium_104 LFIERLRSVGGLPLSLDTTSLR--IGAIR---IDADLDKQDVFALIEQEL
MMAR_4842|M.marinum_M VVRSRRYLANGRPVETAVSYIPAAFAEGTR--IEQVDTGPGGIYARLEEH
MUL_0525|M.ulcerans_Agy99 VVRSRRYLADGKPVETATSYIPVAFAQGTK--IEQTDTGPGGIYARFEEA
MSMEG_6745|M.smegmatis_MC2_155 VQLLRLRLLDGSPAMLERAAFVERIGRTL---FDFDCDSGSIWAFLQSR-
TH_0099|M.thermoresistible__bu VLGYRQADGSDTPVCRTEYYINRAFAAVG---RLLPSHTGPIFPLIEDMF
* . * : .
Mb0816c|M.bovis_AF2122/97 GVRIAQATHEIHAAGASPDVADALGLAVGSPVLVVDRTSYTNDGKPLEVV
Rv0792c|M.tuberculosis_H37Rv GVRIAQATHEIHAAGASPDVADALGLAVGSPVLVVDRTSYTNDGKPLEVV
Mflv_2652|M.gilvum_PYR-GCK AIELARTTAVLESIVLDDASSVQLGVRAGSAGTLLTRTTYDSADRCVEYA
Mvan_3927|M.vanbaalenii_PYR-1 GVELARSTAVLESIVLDDASSMQLGVRAGSAGTLLTRTTVDTAGRCVEYA
MAB_2239|M.abscessus_ATCC_1997 GRRLGSANLALEAVAADSHSAATLQVPDSAPLLLLERLTHLDDGTPVDLE
MAV_5032|M.avium_104 GERLGWAEITVESVLADPNTATLLQIRTGAPLLLLHRLTHLRDGTPFDLE
MMAR_4842|M.marinum_M GHVLARFTEEVAARMPTPEERRQLELDSGVPVLTVLRTAYDTNDVAVEVC
MUL_0525|M.ulcerans_Agy99 GHTLARFNEEVGARMPTPEERRALQLPPGAPVLTVVRTAYDTKDVAVEVC
MSMEG_6745|M.smegmatis_MC2_155 GVQLATASHTIDAVGADPVDATNLQVPEGTPLLRQRRTTRAADGEVIEFH
TH_0099|M.thermoresistible__bu GVSIIEVHQQISAVLLGPGLADRLRDEAGSPALEMQRTYKTSDGDVAQVT
. : : : * . . * . :
Mb0816c|M.bovis_AF2122/97 VFHHRPERYQFSVTLPRTLPGSGAGIIEKRDFA
Rv0792c|M.tuberculosis_H37Rv VFHHRPERYQFSVTLPRTLPGSGAGIIEKRDFA
Mflv_2652|M.gilvum_PYR-GCK RDVYRADRAAFEVSEELRSQRLSAV--------
Mvan_3927|M.vanbaalenii_PYR-1 RDVYRADRASFEVSEELRTPRLSAV--------
MAB_2239|M.abscessus_ATCC_1997 YIRMRGDRITLRSNLLRDI--------------
MAV_5032|M.avium_104 TVRYRGDRCSLVTTATREDAPPQ----------
MMAR_4842|M.marinum_M DTVKVASAYLLEYEFPAR---------------
MUL_0525|M.ulcerans_Agy99 DTVKVASAYLLEYDFSAR---------------
MSMEG_6745|M.smegmatis_MC2_155 DDRYLPGLVTFTLENTLETRTTLARNATR----
TH_0099|M.thermoresistible__bu VNTHPASRFRHSMTMRRVKP-------------