For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAAAEPTPLRRPRADQARLVADVLRHQIHAGGYAEGGLPSEHALAAEFFVSRNTVREALTTLKDEGLIER GPRTGTHVALRKYEHGLDALLGLKETFKGYGEVRNEVRVIQEVNAPPAVAHRLRLDPGARVIFIERLRYL GDLPLSLDKTYLTPDVGEAVIEHQLETNDVFALIERVTGRRLGSANLALEAVAADSHSAATLQVPDSAPL LLLERLTHLDDGTPVDLEYIRMRGDRITLRSNLLRDI
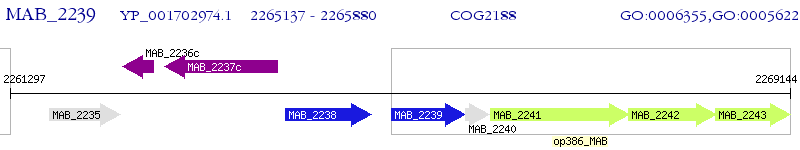
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2239 | - | - | 100% (247) | GntR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_0953 | - | 6e-21 | 31.30% (230) | GntR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_1498c | - | 8e-15 | 30.08% (236) | GntR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0816c | - | 4e-15 | 29.91% (234) | GntR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2652 | - | 9e-15 | 29.85% (201) | regulatory protein GntR, HTH |
| M. tuberculosis H37Rv | Rv0792c | - | 4e-15 | 29.91% (234) | GntR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_2560 | - | 6e-15 | 30.49% (223) | GntR family transcriptional regulator |
| M. avium 104 | MAV_3402 | - | 2e-96 | 73.55% (242) | GntR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_1227 | - | 5e-47 | 48.48% (231) | transcriptional regulator, GntR family protein |
| M. thermoresistible (build 8) | TH_3932 | - | 2e-14 | 29.39% (228) | PUTATIVE transcriptional regulator, GntR family |
| M. ulcerans Agy99 | MUL_3201 | - | 4e-15 | 30.04% (223) | GntR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0056 | - | 3e-18 | 33.18% (214) | UbiC transcription regulator-associated domain-containing |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0816c|M.bovis_AF2122/97 MTS----VKLDLDAADLRISRGSVPASTQLAEALKAQIIQQRLPRGGRLP
Rv0792c|M.tuberculosis_H37Rv MTS----VKLDLDAADLRISRGSVPASTQLAEALKAQIIQQRLPRGGRLP
MMAR_2560|M.marinum_M MTSDEDSPRHDLADADLRVTRGSVPASVQLAELLRTQILERALSPGSRLP
MUL_3201|M.ulcerans_Agy99 MTSDEDSPRHDLADADLRVTRGSVPASVQLAELLRTQILERALSPGSRLP
MAB_2239|M.abscessus_ATCC_1997 ----------MAAAEPTPLRRPRADQARLVADVLRHQIHAGGYAEGG-LP
MAV_3402|M.avium_104 ------------MAHPVPIGGPRADRARRVADVLRQQIHADAYPDG--LP
MSMEG_1227|M.smegmatis_MC2_155 ---------------------------MLRGKILDGVYGGLAAARPM-LP
Mvan_0056|M.vanbaalenii_PYR-1 ---------------------MNVPKPYLVRTGLDGILDG--LQEGDPFP
TH_3932|M.thermoresistible__bu -----MTRTATPMRSTTINRFADRPLYRQLSEVLEARLIER-ARPGDRLP
Mflv_2652|M.gilvum_PYR-GCK -----MSTTEADLRGYSAKMPAGTPLYMSIASDVRDLIASEALGPHTLLP
: :*
Mb0816c|M.bovis_AF2122/97 SERELIDRSGLSRVTVRAAVGMLQRQGWLVRRQGLGTFVADP--VEQELS
Rv0792c|M.tuberculosis_H37Rv SERELIDRSGLSRVTVRAAVGMLQRQGWLVRRQGLGTFVADP--VEQELS
MMAR_2560|M.marinum_M TEQELIARSGVSRSTVRAAVGILEDEGWLVRRQGLGTFVAEP--VKQDLE
MUL_3201|M.ulcerans_Agy99 TEQELIARSGVSRSTVRAAVGILEDEGWLVRRQGLGTFVAEP--VEQDLE
MAB_2239|M.abscessus_ATCC_1997 SEHALAAEFFVSRNTVREALTTLKDEGLIERGPRTGTHVALR--KYEHGL
MAV_3402|M.avium_104 AELELAAEFSVSRNTIREALAVLKREGLIDRGPRVGTHVAQR--KYDHAL
MSMEG_1227|M.smegmatis_MC2_155 PENELAAEFGVSRNAIREALELLRGEGLITRVQGAGTFVTGA--KLSQRI
Mvan_0056|M.vanbaalenii_PYR-1 PERELAARFGVARETVRQALHELLVEGRIERR-GRGTVVSRP--KLVQPL
TH_3932|M.thermoresistible__bu SEAELSGQFGVNRLTVRRALHELAQRGIIETVHGRGSFVARQPVRYRLSA
Mflv_2652|M.gilvum_PYR-GCK SERELAEQHGVSRMTARQALSLLESEGVVYRKPPRGTFVAEP-----RVR
.* * . : * : * *: * .* : *: *:
Mb0816c|M.bovis_AF2122/97 CGVRTITEVLLSCGVTPQVDVLSHQTGPAPQRISETLGLVE---VLCIRR
Rv0792c|M.tuberculosis_H37Rv CGVRTITEVLLSCGVTPQVDVLSHQTGPAPQRISETLGLVE---VLCIRR
MMAR_2560|M.marinum_M SGVRTITEVLLSRGVTPRVEVLSHAVVAAPQSVGAALGRSE---VLRIQR
MUL_3201|M.ulcerans_Agy99 SGVRTITEVLLSRGVTPRVEVLAHAVVAAPQSVGAALGRSE---VLRIQR
MAB_2239|M.abscessus_ATCC_1997 DALLGLKETFKGYG-EVRNEVRVIQEVNAPPAVAHRLRLDPGARVIFIER
MAV_3402|M.avium_104 DALLGLKETFKDLG-EVRNEVRAAMPVTAPPSVARRLRLAPGEQAVFIER
MSMEG_1227|M.smegmatis_MC2_155 DRLEGLAESLAGHQLPVENTVLSVRESVASPFVAAKLQLPEGAPILFIER
Mvan_0056|M.vanbaalenii_PYR-1 S-LRSYTEGAQRMGRTPGRLLVTWENITVDAEIAAALDVSPSARVMHLER
TH_3932|M.thermoresistible__bu TRDASFTRGMRELGHPVRIEVLGAETTRCRR---LRTELHTAGEVLTTTT
Mflv_2652|M.gilvum_PYR-GCK FHIGSFSEEVARMGRRPAAQLLWAEHQAATPAVRRALGLDEGAAVHVFHR
. :
Mb0816c|M.bovis_AF2122/97 RIRTGDQPLALVTAYLPPGVG-PAVEPLLSGSADTETTYAMWERRLGVRI
Rv0792c|M.tuberculosis_H37Rv RIRTGDQPLALVTAYLPPGVG-PAVEPLLSGSADTETTYAMWERRLGVRI
MMAR_2560|M.marinum_M RYSESDQPLAIVTCYLPADLPHDAVEPLLSATSATATTYTMWEQRLGVRI
MUL_3201|M.ulcerans_Agy99 RYSESDQPLAIVTCYLPTDLPHDAVEPLLSATSVTATTYTMWEQRLGVRI
MAB_2239|M.abscessus_ATCC_1997 LRYLGDLPLSLDKTYLTPDVGEAVIEHQLE----TNDVFALIERVTGRRL
MAV_3402|M.avium_104 LRYLGELPLSLDLTYLAPPIGRQVLEHSLE----TNDVFALIEQVSGQRL
MSMEG_1227|M.smegmatis_MC2_155 LRSVGGVPLSLDTTSLR--LGAVSVDDNLD----RQDVFALIERELGMRL
Mvan_0056|M.vanbaalenii_PYR-1 VLLADDVRIGLESTYLPRHRFGDLEQHFDP----TTSLYSAIRAR-GVKF
TH_3932|M.thermoresistible__bu LRHVDGQPWSVSVTSIALDRFPGIAQQWSG----STSLFDFLWQTFGVRM
Mflv_2652|M.gilvum_PYR-GCK LRSVDEVPFALETTFLPAELTPGILDMTED-----GSLWAVLRSKYAIEL
. .: : : : . .:
Mb0816c|M.bovis_AF2122/97 AQATHEIHAAGASPDVADALGLAVGSPVLVVDRTSYTNDGKPLEVVVFHH
Rv0792c|M.tuberculosis_H37Rv AQATHEIHAAGASPDVADALGLAVGSPVLVVDRTSYTNDGKPLEVVVFHH
MMAR_2560|M.marinum_M SSATHEIQAAGASPEIAAALGLSEGAPVLVLRRTSYTDGDKPLEVVLFHH
MUL_3201|M.ulcerans_Agy99 SSATHEVQAAGASPEIASALGLSEGAPVLVLRRTSYTDGDKPLEVVLFHH
MAB_2239|M.abscessus_ATCC_1997 GSANLALEAVAADSHSAATLQVPDSAPLLLLERLTHLDDGTPVDLEYIRM
MAV_3402|M.avium_104 GSAALALEAIPADAHSAATLQVPDGAALLMLERLTSLADGTPVDLEYIRM
MSMEG_1227|M.smegmatis_MC2_155 GWAEMTVESVSADADTAKLLQIRSGAPLLLLHRLTHLEDGTPFDFETVRY
Mvan_0056|M.vanbaalenii_PYR-1 GSATERIETVLPSPREAELLESTTAMPMLLLNRRSVDVDGVPIEVVRALY
TH_3932|M.thermoresistible__bu RRAYRSFSAVLADLAEAQHLGLRTGAPVLEMRGLNVDQHGAPVAVVRHRF
Mflv_2652|M.gilvum_PYR-GCK ARTTAVLESIVLDDASSVQLGVRAGSAGTLLTRTTYDSADRCVEYARDVY
: . : . : * . . : . . .
Mb0816c|M.bovis_AF2122/97 RPERYQFSVTLPRTLPGSGAGIIEKRDFA
Rv0792c|M.tuberculosis_H37Rv RPERYQFSVTLPRTLPGSGAGIIEKRDFA
MMAR_2560|M.marinum_M RPERYEFCVTLPRTMPGQRAGMTER----
MUL_3201|M.ulcerans_Agy99 RPERYEFCVTLPRTMPGQRAGMTER----
MAB_2239|M.abscessus_ATCC_1997 RGDRITLRSNLLRDI--------------
MAV_3402|M.avium_104 RGDRITMRGNLFRAEPMRSNS--------
MSMEG_1227|M.smegmatis_MC2_155 RGDRCTLMTTAARGVDDPPQAPQEPPQ--
Mvan_0056|M.vanbaalenii_PYR-1 RGDRVAFEATLDDR---------------
TH_3932|M.thermoresistible__bu RGDRVELTVDLA-----------------
Mflv_2652|M.gilvum_PYR-GCK RADRAAFEVSEELRSQRLSAV--------
* :* :