For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTATLDYDGDIAVLDLGDDENRFTPQFLDEVDALLDAVLEKGAHGLVTTADSKFYSNGLDLEWLGAHSDQ GAWYVGRVQGLLARVLTFPLPTAAAVVGHAFGAGAMLAMAHDYRVMREDRGFFCFPEVDIHIPFTPGMAA LIQAKLTPQAAIASMTTGRRFGGSEAHEHGIVDATAHEGAVTLAATSLLRPLGSKDQGTLGAIKQVMFGA AAAALRNP
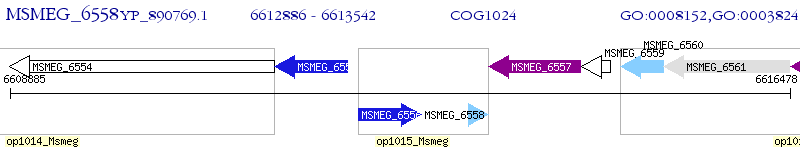
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6558 | - | - | 100% (218) | putative enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_4775 | - | 6e-13 | 30.91% (220) | enoyl-CoA hydratase/isomerase family protein |
| M. smegmatis MC2 155 | MSMEG_4077 | - | 5e-11 | 24.29% (210) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2511 | echA14 | 1e-10 | 30.65% (199) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2522 | - | 4e-11 | 28.64% (213) | enoyl-CoA hydratase/isomerase |
| M. tuberculosis H37Rv | Rv2486 | echA14 | 1e-10 | 30.65% (199) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02498 | - | 3e-89 | 74.42% (215) | putative enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_2465c | - | 7e-73 | 63.26% (215) | enoyl-coa hydratase/isomerase family protein |
| M. marinum M | MMAR_4317 | echA1_1 | 4e-10 | 28.82% (170) | enoyl-CoA hydratase, EchA1_1 |
| M. avium 104 | MAV_0967 | - | 1e-11 | 30.00% (200) | enoyl-CoA hydratase/isomerase family protein |
| M. thermoresistible (build 8) | TH_2890 | - | 1e-12 | 32.18% (174) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_0713 | echA3 | 4e-09 | 24.88% (217) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_4083 | - | 3e-12 | 34.32% (169) | enoyl-CoA hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6558|M.smegmatis_MC2_155 --------MTATLDYDGDIAVLDLGDDENR--FTPQFLDEVDALLDAVLE
MLBr_02498|M.leprae_Br4923 --------MTIAVHYDEKIAILELGDDENR--FTSVFLEEINSHLDEIIA
MAB_2465c|M.abscessus_ATCC_199 --------MTPTLELQDTIAVLNLGADENR--FSPDWLETVDGLLDDVLT
Mb2511|M.bovis_AF2122/97 ----MAQYDPVLLSVDKHVALITVNDPDRRNAVTDEMSAQLRAAIQRAEG
Rv2486|M.tuberculosis_H37Rv ----MAQYDPVLLSVDKHVALITVNDPDRRNAVTDEMSAQLRAAIQRAEG
Mvan_4083|M.vanbaalenii_PYR-1 ------MSELVLTQVEDRVALITINDPDRRNAVTAEISAALRAAVEAAEA
Mflv_2522|M.gilvum_PYR-GCK ---------MVDLEIDDGLAIITIDRPHARNAISLETMDQLDTALDGAHG
MAV_0967|M.avium_104 ---------MVDLEIDDGLAVLTIDRPHARNAIALDTMDQLEKALDAAAG
TH_2890|M.thermoresistible__bu ---VTADAPLVLREDRGAVALLWLNRAAQRNALRYESWTELSEHLAVLAE
MMAR_4317|M.marinum_M MTDAGDDQPAGLVEHRGNVMVITINRPEARNAINGAVSVAVGDALESAQN
MUL_0713|M.ulcerans_Agy99 ------MSGPVTYTHDDAIGVIRMDD-GKVNVLGPTMQQALNEAIDAADR
: :: :. . : :
MSMEG_6558|M.smegmatis_MC2_155 KG-AHGLVTTADS-KFYSNGLDLEWLGAHS---DQGAWYVGRVQGLLARV
MLBr_02498|M.leprae_Br4923 SG-ANGLVTTALG-KFYSNGLDLDWLSAHR---DQTQWYVGKVQGLLARL
MAB_2465c|M.abscessus_ATCC_199 Q--AQALVTVGTG-KFYSNGLDLDWLMSHG---DRTDWYVGRVHALFRRV
Mb2511|M.bovis_AF2122/97 DPDVHAVVVTGAG-KAFCAGADLSALGAGVG-DPAEPRLLRLYDGFMA-V
Rv2486|M.tuberculosis_H37Rv DPDVHAVVVTGAG-KAFCAGADLSALGAGVG-DPAEPRLLRLYDGFMA-V
Mvan_4083|M.vanbaalenii_PYR-1 DTGVHAVIVTGAG-KAFCAGADLTALGAAT-----EDGLRVIYDGFLA-V
Mflv_2522|M.gilvum_PYR-GCK ---ARALALTGAGDKAFVSGGDLKELSALRT-VEQAASMSFRMRSICDRI
MAV_0967|M.avium_104 ---AQALVITGAGDRAFVSGGDLKELSALRT-EEDAAAMARRMRAICDQL
TH_2890|M.thermoresistible__bu DPDVRGVXVAGAG-GFFSAGGDLKTGPAHGAGPLGPAGRVEHAQRVMEQL
MMAR_4317|M.marinum_M DPEVRAVVITGAGDKSFCAGADLLAISRREN-IYHPDHNEWGFAGYVHHF
MUL_0713|M.ulcerans_Agy99 DN-VGALVIAGNH-RVFSGGFDLKVLTSGEA--KPAIDMLRGGFELSYRL
. .: .. : * ** .
MSMEG_6558|M.smegmatis_MC2_155 LTFPLPTAAAVVGHAFGAGAMLAMAHDYRVMREDRGFFCFPEVDIHIPFT
MLBr_02498|M.leprae_Br4923 LTLPIPTAAAVVGHAFGAGAMLAMAHDFRVMRADRGYFCFPEVDIRIPFT
MAB_2465c|M.abscessus_ATCC_199 LTFGLPTVAAINGHAFGAGAMLAVAHDYRVMRSDRGFLCFPEVDIHIPFT
Mb2511|M.bovis_AF2122/97 SSCNLPTIAAVNGAAVGAGLNLALAADVRIAG-PAALFDARFQKLGLHPG
Rv2486|M.tuberculosis_H37Rv SSCNLPTIAAVNGAAVGAGLNLALAADVRIAG-PAALFDARFQKLGLHPG
Mvan_4083|M.vanbaalenii_PYR-1 AECNLPTIAAVNGPAVGAGLNLALAADVRIAG-PAALFDPRFQKLGIHPG
Mflv_2522|M.gilvum_PYR-GCK AGFPAPVVAALNGHALGGGAEFAVAADIRLAA-DDIKIGFNQVALEIMPA
MAV_0967|M.avium_104 ADFPAPVVAALNGHAFGGGAEVAVAADIRVAA-DDIKIAFNQVALEIMPA
TH_2890|M.thermoresistible__bu KAVPVPTVAAVEGAAVGLGWSLALACDLVVAA-SDAFFSAPFVARAVVPD
MMAR_4317|M.marinum_M IDK--PTIAAVNGTALGGGTELALASDLVVAD-ERAKFGLPEVKRGLIAA
MUL_0713|M.ulcerans_Agy99 LSYPKPVVIACTGHAIAMGAFLLCSGDHRVAA-HAYNIQANEVAIGMTIP
*. * * *.. * . : * : : :
MSMEG_6558|M.smegmatis_MC2_155 PGMAALIQAKLTPQAAIASMTTGRRFGGSEAHEHGIVDATAHEGAVTLAA
MLBr_02498|M.leprae_Br4923 PGMAALIQAKLAPQAATASMTTGRRFGGGEAARLGIVDNTAAEDAVTAVA
MAB_2465c|M.abscessus_ATCC_199 PGMASLIQAKVGPQTAVTAMTTGHRYGGAAAVAAGLADSCAAEHEVLSTA
Mb2511|M.bovis_AF2122/97 GGATWMLQRAVGPQVARAALLFGMCFDAESAVRHGLALMVADDPVTAALE
Rv2486|M.tuberculosis_H37Rv GGATWMLQRAVGPQVARAALLFGMCFDAESAVRHGLALMVADDPVTAALE
Mvan_4083|M.vanbaalenii_PYR-1 GGATWMLQRAIGPQAARAALLFGMRFDAESAVRHGLALEVADDPVAAARE
Mflv_2522|M.gilvum_PYR-GCK WGGAERLVELVGYSRALLLAGTGRILAATDAERLGLIDQVIPRASFD--E
MAV_0967|M.avium_104 WGGAERLAALVGQSRALLLAGSGTALDAVEAERIGLVDRVLPRDVFDSAE
TH_2890|M.thermoresistible__bu GGLAWRLTHQLGRQRASALLLRGSRLAAPDAAQAGLVTELAELGSAVEVA
MMAR_4317|M.marinum_M AGGVFRIVNQLPRKVAMEMLLTGEPITASDAYEWGLINQVVKQGAVLDAA
MUL_0713|M.ulcerans_Agy99 YAAMEVLKLRLTPSAYQQAAGLAKTFFGETALAAGFIDEISLPEVVLSRA
. : : . . . * *:
MSMEG_6558|M.smegmatis_MC2_155 TSLLRPLGSKDQGTLGAIKQVMFGAAAAALRNP-----------------
MLBr_02498|M.leprae_Br4923 VDLLHPLGGKDPGTLGVIKQTMFGHAAAALSANA----------------
MAB_2465c|M.abscessus_ATCC_199 IDLMKPLTGKDSGTLSAIKATMFAATATALAA------------------
Mb2511|M.bovis_AF2122/97 LAAGPAAAPREVVLASKAT--MRATASPGSLDLEQHELAKRLELGPQAKS
Rv2486|M.tuberculosis_H37Rv LAAGPAAAPREVVLASKAT--MRATASPGSLDLEQHELAKRLELGPQAKS
Mvan_4083|M.vanbaalenii_PYR-1 LAAGPASAPRDVVVSTKAS--MRATYNPGISDLHQHHMAVDIEIGPQARS
Mflv_2522|M.gilvum_PYR-GCK Q-WRLTARLLASVPAGEVKRVMRGVPTTEAVAAFARLWVSDEHWAAADKV
MAV_0967|M.avium_104 QGWRSIARSLARRPAAEIKRVIRGVSPEEAIASFARLWVADPHWQAAERV
TH_2890|M.thermoresistible__bu LALLDELASADAGAVEMTKRLINSVEAAQLAAFHPLELAMATVAQQRSAS
MMAR_4317|M.marinum_M LALAARVTVNAPLSVRASKRIAYGVDDGVVADEERGWERTMREMRSLIRS
MUL_0713|M.ulcerans_Agy99 EEAAREFAGLNQQAHNATKLRARAEALKAIRAGIDGIEAEFGL-------
. .
MSMEG_6558|M.smegmatis_MC2_155 ---------------------
MLBr_02498|M.leprae_Br4923 ---------------------
MAB_2465c|M.abscessus_ATCC_199 ---------------------
Mb2511|M.bovis_AF2122/97 VQSPEFAARLAAAQHR-----
Rv2486|M.tuberculosis_H37Rv VQSPEFAARLAAAQHR-----
Mvan_4083|M.vanbaalenii_PYR-1 IESPEFAARLAAAQRR-----
Mflv_2522|M.gilvum_PYR-GCK MKKGK----------------
MAV_0967|M.avium_104 MARNTKPRPAAGGAS------
TH_2890|M.thermoresistible__bu AAARDRW--------------
MMAR_4317|M.marinum_M EDAKEGPLAFAQKREPVWKAR
MUL_0713|M.ulcerans_Agy99 ---------------------