For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDLEIDDGLAIITIDRPHARNAISLETMDQLDTALDGAHGARALALTGAGDKAFVSGGDLKELSALRTVE QAASMSFRMRSICDRIAGFPAPVVAALNGHALGGGAEFAVAADIRLAADDIKIGFNQVALEIMPAWGGAE RLVELVGYSRALLLAGTGRILAATDAERLGLIDQVIPRASFDEQWRLTARLLASVPAGEVKRVMRGVPTT EAVAAFARLWVSDEHWAAADKVMKKGK*
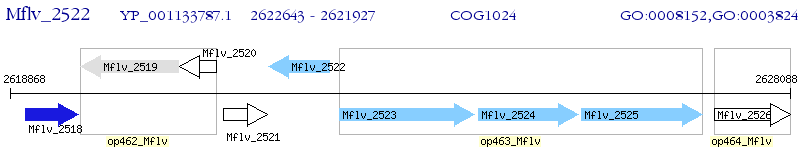
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2522 | - | - | 100% (238) | enoyl-CoA hydratase/isomerase |
| M. gilvum PYR-GCK | Mflv_2036 | - | 5e-30 | 36.27% (193) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2117 | - | 6e-24 | 37.63% (194) | enoyl-CoA hydratase/isomerase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1099c | echA8 | 1e-28 | 33.17% (208) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv1070c | echA8 | 1e-28 | 33.17% (208) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 1e-25 | 35.67% (171) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_1185c | - | 3e-28 | 33.33% (192) | enoyl-CoA hydratase |
| M. marinum M | MMAR_4691 | echA8_2 | 6e-90 | 69.83% (242) | enoyl-CoA hydratase, EchA8_2 |
| M. avium 104 | MAV_0967 | - | 2e-95 | 72.80% (239) | enoyl-CoA hydratase/isomerase family protein |
| M. smegmatis MC2 155 | MSMEG_4775 | - | 1e-105 | 78.15% (238) | enoyl-CoA hydratase/isomerase family protein |
| M. thermoresistible (build 8) | TH_3655 | - | 3e-81 | 71.43% (210) | PUTATIVE short chain enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_0310 | echA8_2 | 1e-89 | 69.83% (242) | enoyl-CoA hydratase, EchA8_2 |
| M. vanbaalenii PYR-1 | Mvan_4136 | - | 1e-120 | 90.76% (238) | enoyl-CoA hydratase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2522|M.gilvum_PYR-GCK -----------------MVDLEID---DGLAIITIDRPHARNAISLETMD
Mvan_4136|M.vanbaalenii_PYR-1 MVFSSCENANTVNGECPMVDLELD---DGLAVITIDRPHARNAISLETME
MSMEG_4775|M.smegmatis_MC2_155 -------------MENDLVDLELD---GELAVITIDRPHARNAISLDTMD
MMAR_4691|M.marinum_M -----------------MVDLEFDELDTGLAVLTIDRPHARNAIGLETMA
MUL_0310|M.ulcerans_Agy99 -----------------MVDLEFDGLDTGLAVLTIDRPHARNAIGLETMA
MAV_0967|M.avium_104 -----------------MVDLEID---DGLAVLTIDRPHARNAIALDTMD
TH_3655|M.thermoresistible__bu -----------------MVDLELT---DGLAVITIDRPQARNAIAPATME
Mb1099c|M.bovis_AF2122/97 ----------------MTYETILVERDQRVGIITLNRPQALNALNSQVMN
Rv1070c|M.tuberculosis_H37Rv ----------------MTYETILVERDQRVGIITLNRPQALNALNSQVMN
MLBr_02402|M.leprae_Br4923 ----------------MKYDTILVDGYQRVGIITLNRPQALNALNSQMMN
MAB_1185c|M.abscessus_ATCC_199 -------------MTSHNFETILTERIDRVAVITLNRPKALNALNSQVMN
: : :.::*::**:* **: *
Mflv_2522|M.gilvum_PYR-GCK QLDTALDGAHG---ARALALTGAGDKAFVSGGDLKELSALRTVEQAASMS
Mvan_4136|M.vanbaalenii_PYR-1 QLDKALDGAEG---AAALALTGAGDKAFVSGGDLKELSALRTEEQAAAMA
MSMEG_4775|M.smegmatis_MC2_155 ELDKALDGAAG---SRALAITGAGDRAFVSGGDLKELAALRTEREAGAMA
MMAR_4691|M.marinum_M QLQDALDAAAG---AQALVIKGAGDRAFVSGGDLKELSALRTLEDASAMA
MUL_0310|M.ulcerans_Agy99 QLQDALDAAAG---AQALVIKGAGDRAFVSGGDLKELSALRTLEDASAMA
MAV_0967|M.avium_104 QLEKALDAAAG---AQALVITGAGDRAFVSGGDLKELSALRTEEDAAAMA
TH_3655|M.thermoresistible__bu QLEKALDSLAGNPDAQALVIRGAGDRAFVSGGDLKELAALRTELEASAMA
Mb1099c|M.bovis_AF2122/97 EVTSAATELDDDPDIGAIIITGS-AKAFAAGADIKEMADL---TFADAFT
Rv1070c|M.tuberculosis_H37Rv EVTSAATELDDDPDIGAIIITGS-AKAFAAGADIKEMADL---TFADAFT
MLBr_02402|M.leprae_Br4923 EITNAAKELDIDPDVGAILITGS-PKVFAAGADIKEMASL---TFTDAFD
MAB_1185c|M.abscessus_ATCC_199 EVTTAAAEFDADHGIGAIIITGS-EKAFAAGADIKEMSEQ---SFSDMFG
:: * *: : *: :.*.:*.*:**:: : :
Mflv_2522|M.gilvum_PYR-GCK FRMRSICDRIAGFPAPVVAALNGHALGGGAEFAVAADIRLAADDIKIGFN
Mvan_4136|M.vanbaalenii_PYR-1 FRMRSICDRIAGFPGPVLAALNGHALGGGAEFAVSADIRVAADDIKIGFN
MSMEG_4775|M.smegmatis_MC2_155 WRMRSICDRIAGFPGPVIAALNGHALGGGAEVAVSADIRIAADDVKIGFN
MMAR_4691|M.marinum_M KRMRSICDQIAAFPAPVIAALNGHAFGGGAEVAVAADIRVAADDIKIAFN
MUL_0310|M.ulcerans_Agy99 KRMRSICDQIATFPAPVIAALNGHAFGGGAEVAVAADIRVAADDIKIAFN
MAV_0967|M.avium_104 RRMRAICDQLADFPAPVVAALNGHAFGGGAEVAVAADIRVAADDIKIAFN
TH_3655|M.thermoresistible__bu WRMRSICDRLARFPGPVIAALNGHALGGGAEVAVAADIRIAADDIRIGFN
Mb1099c|M.bovis_AF2122/97 ADFFATWGKLAAVRTPTIAAVAGYALGGGCELAMMCDVLIAADTAKFGQP
Rv1070c|M.tuberculosis_H37Rv ADFFATWGKLAAVRTPTIAAVAGYALGGGCELAMMCDVLIAADTAKFGQP
MLBr_02402|M.leprae_Br4923 ADFFSAWGKLAAVRTPMIAAVAGYALGGGCELAMMCDLLIAADTAKFGQP
MAB_1185c|M.abscessus_ATCC_199 SDFFSAWGKLGAVRTPTIAAVSGYALGGGCELAMMCDVIIAAENATFGQP
: : .::. . * :**: *:*:***.*.*: .*: :**: :.
Mflv_2522|M.gilvum_PYR-GCK QVALEIMPAWGGAERLVELVGYSRALLLAGTGRILAATDAERLGLIDQVI
Mvan_4136|M.vanbaalenii_PYR-1 QVALAIMPAWGGAERLVKLVGYSRALLLAGTGRILDAADAERIGLIDQVI
MSMEG_4775|M.smegmatis_MC2_155 QVALEIMPAWGGAERLVTLVGRSRALLLAGTGRVLDAAEAERVGLVDQVL
MMAR_4691|M.marinum_M QVLLEIMPAWGGAERLAAVVGKSKALLLAGSGIALDAVEAERIGLVDLVF
MUL_0310|M.ulcerans_Agy99 QVLLEIMPAWGGAERLAAVVGKSKALLLAGSGIALDAVEAERIGLVDLVF
MAV_0967|M.avium_104 QVALEIMPAWGGAERLAALVGQSRALLLAGSGTALDAVEAERIGLVDRVL
TH_3655|M.thermoresistible__bu QVALAIMPAWGGAERLAELVGRSKALLLAGTGTVLTAAEAERVGLVDKVV
Mb1099c|M.bovis_AF2122/97 EIKLGVLPGMGGSQRLTRAIGKAKAMDLILTGRTMDAAEAERSGLVSRVV
Rv1070c|M.tuberculosis_H37Rv EIKLGVLPGMGGSQRLTRAIGKAKAMDLILTGRTMDAAEAERSGLVSRVV
MLBr_02402|M.leprae_Br4923 EIKLGVLPGMGGSQRLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVV
MAB_1185c|M.abscessus_ATCC_199 EIKLGVLPGMGGSQRLTRAIGKAKAMDMILTGRNMDAAEAERSGLVSRVV
:: * ::*. **::**. :* ::*: : :* : *.:*** **:. *.
Mflv_2522|M.gilvum_PYR-GCK PRASFD---EQWRLTARLLASVPAGEVKR--------------VMRGVPT
Mvan_4136|M.vanbaalenii_PYR-1 PRASFD---EQWRLIARSLASAPAGEVKR--------------VMRGVPT
MSMEG_4775|M.smegmatis_MC2_155 PRAEFA---DGWRALAHTLAGRPAGEIKR--------------VMGGVPA
MMAR_4691|M.marinum_M PRASFDSTDGGWRSLARSLARRPAAEVKR--------------VINGTSP
MUL_0310|M.ulcerans_Agy99 PRASFDSTDGGWRSLARSLARRPAAEVKR--------------VINGTSP
MAV_0967|M.avium_104 PRDVFDSAEQGWRSIARSLARRPAAEIKR--------------VIRGVSP
TH_3655|M.thermoresistible__bu PRASFD---TEWRAIARSLATRPAAEIKRGARGGRRQPDEPDEVAVGFSP
Mb1099c|M.bovis_AF2122/97 PADDLLTEARATATTISQMSASAARMAKEA-------------VNRAFES
Rv1070c|M.tuberculosis_H37Rv PADDLLTEARATATTISQMSASAARMAKEA-------------VNRAFES
MLBr_02402|M.leprae_Br4923 LADDLLPEAKAVATTISQMSRSATRMAKEA-------------VNRSFES
MAB_1185c|M.abscessus_ATCC_199 ATESLLDEAKAVAKTISEMSLSASMMAKEA-------------VNRAFES
: :: .: *. * . .
Mflv_2522|M.gilvum_PYR-GCK TEAVAAFARLWVSDEHWAAADKVMKKGK----------------------
Mvan_4136|M.vanbaalenii_PYR-1 TEAVAAFARLWASDEHWAAADKVMKKGK----------------------
MSMEG_4775|M.smegmatis_MC2_155 TEAVAAFARLWVAEEHWAAADKVMNKKK----------------------
MMAR_4691|M.marinum_M DEAIASFARLWVEDAHWQAAERVMNRTPNRGPGAEGAT------------
MUL_0310|M.ulcerans_Agy99 DEAIASFARLWVEDAHWQAAEQVMNRTPNRGPGAEGAT------------
MAV_0967|M.avium_104 EEAIASFARLWVADPHWQAAERVMARNTKPRPAAGGAS------------
TH_3655|M.thermoresistible__bu GSRIARSDRRPHNRSAGSAREVPSLRAQLRGQVPTGEQPRGRGHPAPDPP
Mb1099c|M.bovis_AF2122/97 SLSEGLLYERRLFHSAFATEDQSEGMAAFIEKRAPQFTHR----------
Rv1070c|M.tuberculosis_H37Rv SLSEGLLYERRLFHSAFATEDQSEGMAAFIEKRAPQFTHR----------
MLBr_02402|M.leprae_Br4923 TLAEGLLHERRLFHSTFVTDDQSEGMAAFIEKRAPQFTHR----------
MAB_1185c|M.abscessus_ATCC_199 SLAEGLLFERRIFHSAFGTADQSEGMAAFVEKRPANFIHR----------
. . : :
Mflv_2522|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4136|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4775|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4691|M.marinum_M --------------------------------------------------
MUL_0310|M.ulcerans_Agy99 --------------------------------------------------
MAV_0967|M.avium_104 --------------------------------------------------
TH_3655|M.thermoresistible__bu PVAGAYQLIVVVTDRVDPVLPVVRAQHQHRVHIRARGGPVAVDVDDDVAH
Mb1099c|M.bovis_AF2122/97 --------------------------------------------------
Rv1070c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02402|M.leprae_Br4923 --------------------------------------------------
MAB_1185c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2522|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4136|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4775|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4691|M.marinum_M --------------------------------------------------
MUL_0310|M.ulcerans_Agy99 --------------------------------------------------
MAV_0967|M.avium_104 --------------------------------------------------
TH_3655|M.thermoresistible__bu AVVGRSHRSVHLRHHLGEGVRIGEGAELVVVDVDPVGPAAAEQRPVLLVD
Mb1099c|M.bovis_AF2122/97 --------------------------------------------------
Rv1070c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02402|M.leprae_Br4923 --------------------------------------------------
MAB_1185c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2522|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4136|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4775|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_4691|M.marinum_M --------------------------------------------------
MUL_0310|M.ulcerans_Agy99 --------------------------------------------------
MAV_0967|M.avium_104 --------------------------------------------------
TH_3655|M.thermoresistible__bu TRCVADQYVGDLRPVGVHLSHGNRRGGSRRDHHRPCNRFSLDENVVSGYC
Mb1099c|M.bovis_AF2122/97 --------------------------------------------------
Rv1070c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02402|M.leprae_Br4923 --------------------------------------------------
MAB_1185c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_2522|M.gilvum_PYR-GCK -----------------------------
Mvan_4136|M.vanbaalenii_PYR-1 -----------------------------
MSMEG_4775|M.smegmatis_MC2_155 -----------------------------
MMAR_4691|M.marinum_M -----------------------------
MUL_0310|M.ulcerans_Agy99 -----------------------------
MAV_0967|M.avium_104 -----------------------------
TH_3655|M.thermoresistible__bu HRRRHRARPPGPGRRGPAGGVNAVVQTPG
Mb1099c|M.bovis_AF2122/97 -----------------------------
Rv1070c|M.tuberculosis_H37Rv -----------------------------
MLBr_02402|M.leprae_Br4923 -----------------------------
MAB_1185c|M.abscessus_ATCC_199 -----------------------------