For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTSRANSGARDLELHTVITPGSRTAREDLVTALRDGIRSGRLGTGTVLPPSRVLAADLGLARNTVAEAYA DLVAEGWLASRQGAGTWVARTEAPRLTPPPLRIAGTPKHNLMPGSPDVAEFPRTAWLASARRALSNAPAS ALRMGDPRGRLELRSALAEYLGRVRGVRTTSESIVICSGVRNGVELLGKVFGTQRSIAVEAYGLFIFRDA LAAMGIATAPIGLDEHGAVISDLDMLDVPAVLLTPAHHSPHGMPLHPSRRSEVIEWARRTGGYVIDDDYD GEFRYDRQPVGALQSLDPEHVAYLGSASKSLAQTLRVGWMALPQALVEPVIAAAGGQQFNVDAITALTLA DFIATGGYDRHIRRMRNRYRRRRDHLVEALSGFDVGIRGLAAGVNMLLTLPDGAEPEVLRRAGEAGIALS GLALMRHPLAHPDIPAPDGIIVGFGAPAEHAFRPAVEALRGVLEDVLSP
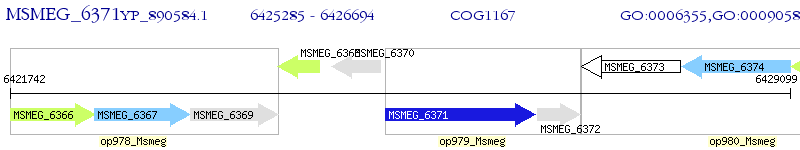
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6371 | - | - | 100% (469) | GntR-family protein regulator |
| M. smegmatis MC2 155 | MSMEG_4140 | - | e-120 | 50.11% (473) | GntR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_5760 | - | e-106 | 47.26% (457) | GntR-family protein regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1211 | - | 6e-08 | 23.45% (307) | N-succinyldiaminopimelate aminotransferase |
| M. gilvum PYR-GCK | Mflv_1181 | - | 1e-180 | 68.22% (472) | GntR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1178 | - | 6e-08 | 23.45% (307) | N-succinyldiaminopimelate aminotransferase |
| M. leprae Br4923 | MLBr_01488 | - | 5e-09 | 24.91% (289) | N-succinyldiaminopimelate aminotransferase |
| M. abscessus ATCC 19977 | MAB_0196c | - | 1e-173 | 65.08% (461) | GntR family transcriptional regulator |
| M. marinum M | MMAR_2376 | - | 8e-20 | 27.05% (451) | GntR family transcriptional regulator |
| M. avium 104 | MAV_3214 | - | 4e-18 | 28.25% (400) | GntR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_4188 | - | 2e-07 | 44.78% (67) | PUTATIVE transcriptional regulator, GntR family |
| M. ulcerans Agy99 | MUL_1552 | - | 3e-20 | 26.95% (449) | GntR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5625 | - | 0.0 | 71.73% (467) | GntR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1211|M.bovis_AF2122/97 --------------------------------------------------
Rv1178|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01488|M.leprae_Br4923 ---------------------------------------------MSPFG
Mflv_1181|M.gilvum_PYR-GCK MVSSWSNSG-------SRDLHLDLREALRPGARGNRNALVAALREAVRSG
Mvan_5625|M.vanbaalenii_PYR-1 -MTSWANSA-------TRDLHLDLRDALVPGARGTKDALISALRDAARSG
MSMEG_6371|M.smegmatis_MC2_155 -MTSRANSG-------ARDLELHT--VITPGSRTAREDLVTALRDGIRSG
MAB_0196c|M.abscessus_ATCC_199 -MSSWANSAKAESRPLSHDLHLDLRQSITPGTRGVRELLTSALRDAVRSG
MMAR_2376|M.marinum_M ------------------------MPVQYRIAGGGAESIAASIESAISQG
MUL_1552|M.ulcerans_Agy99 ------------------------MPVQNRIAGGGAGSIAASIESAISQG
MAV_3214|M.avium_104 ------------------------MAVQKSITGTGAESIAASIEAAISAG
TH_4188|M.thermoresistible__bu -----------------------------VRPRPAYQVLRDRLRDDIAAG
Mb1211|M.bovis_AF2122/97 -MSASLPVFPWDTLAD---------AKALAGAHPDGIVDLSVGT------
Rv1178|M.tuberculosis_H37Rv -MSASLPVFPWDTLAD---------AKALAGAHPDGIVDLSVGT------
MLBr_01488|M.leprae_Br4923 RVSATLPVFPWDTLAD---------AKAVAESHPDGIVDLSVGT------
Mflv_1181|M.gilvum_PYR-GCK RLSAGTTLPPSRALAADLGLARNTVADAYAELVAEGWLASRQGAGTWVVD
Mvan_5625|M.vanbaalenii_PYR-1 RLTAGTMLPSSRTLATDLGLARNTVAEAYAELVAEGWLASRQGAGTWVAD
MSMEG_6371|M.smegmatis_MC2_155 RLGTGTVLPPSRVLAADLGLARNTVAEAYADLVAEGWLASRQGAGTWVAR
MAB_0196c|M.abscessus_ATCC_199 RLTVGSMLPPSRTLASDLGLARNTVAEAYAELVAEGWLESRQGSGTRVVN
MMAR_2376|M.marinum_M GLAPGDALPSIREVAGQLGVNPNTVAAAYRLLRDRGTVETAGRRGTRVRD
MUL_1552|M.ulcerans_Agy99 GLAPGDALPSIREVAGQLGVNPNTVAAAYRLLRDRGTVETAGRRGTRVRD
MAV_3214|M.avium_104 SLAPGDALPPVRELAARLGVNANTAAAAYRLLRHRGAVETAGRRGTRVRH
TH_4188|M.thermoresistible__bu RYRDGTRLPTESELMAQYGVSRQTVRRAFQDLVAEGSVYRVPGR------
: . : * * :
Mb1211|M.bovis_AF2122/97 --------------PVDPVAPL-IQEALAAASAAPGYPAT----------
Rv1178|M.tuberculosis_H37Rv --------------PVDPVAPL-IQEALAAASAAPGYPAT----------
MLBr_01488|M.leprae_Br4923 --------------PVDPVAPL-IQEALAAASSVAGYPTT----------
Mflv_1181|M.gilvum_PYR-GCK TARASSQPPPVPPRPHGARVAP-VHNLMPGSPDVAEFPRSQWAASARRAL
Mvan_5625|M.vanbaalenii_PYR-1 VT----TPAHTPPRPHGVRVAP-THNLMPGSPDVAEFPRSQWLASTRRAL
MSMEG_6371|M.smegmatis_MC2_155 TE----APRLTPP-PLRIAGTP-KHNLMPGSPDVAEFPRTAWLASARRAL
MAB_0196c|M.abscessus_ATCC_199 RG-----KDQLPPRPRGAGTPP-PHNLMPGSGDVTAFPRADWLASARRAL
MMAR_2376|M.marinum_M RP------ASTPRSSLDLGVPAGARDLSTGNPAPELLPLAAVG-------
MUL_1552|M.ulcerans_Agy99 RP------ASTPRSSLDLGVPAGARDLSTGNPAPELLPLAAVG-------
MAV_3214|M.avium_104 RP------ATTPRSLLGLDVPAGVRDLSTGNPHPALLPLAAAAP------
TH_4188|M.thermoresistible__bu --------------------------------------------------
Mb1211|M.bovis_AF2122/97 -------------AGTARLRESVVAALARRYGITRLTEAAVLPVIGTKEL
Rv1178|M.tuberculosis_H37Rv -------------AGTARLRESVVAALARRYGITRLTEAAVLPVIGTKEL
MLBr_01488|M.leprae_Br4923 -------------AGTARLREAAVAALDRRYGITGLTEAAVLPVIGTKEL
Mflv_1181|M.gilvum_PYR-GCK THAPTDAFRMGDPRGRPELRSALAEYLARARGVR-TSPESLVICAGVRHA
Mvan_5625|M.vanbaalenii_PYR-1 GGAPTEALRMGDPRGRPELRSALAEYLARARGVR-TSPESIVICAGVRHA
MSMEG_6371|M.smegmatis_MC2_155 SNAPASALRMGDPRGRLELRSALAEYLGRVRGVR-TTSESIVICSGVRNG
MAB_0196c|M.abscessus_ATCC_199 NHAPHEALRVGDPRGRHELRSAVAEYVSRVRGVR-TTPDSILICAGTRHA
MMAR_2376|M.marinum_M ---WPDGSVLPLLYGAPAMSAELVDYARAALAADGVAADHLAVTNGALDG
MUL_1552|M.ulcerans_Agy99 ---WPDGSVLPLLYGAPAMSAELVDYARAALAADGVAADHLAVTNGALDG
MAV_3214|M.avium_104 ---ARPASGRAVLYGEPAISPALAEYARAALSADGVPADHLALTSGALDG
TH_4188|M.thermoresistible__bu --------------GTYAADRYLRQFGSIEDLMALSDDTTMEVRSGLRRR
* : *
Mb1211|M.bovis_AF2122/97 IA-WLPTLLG-----LGGADLVVVPELAYPTYDVGARLAGTRVLR---AD
Rv1178|M.tuberculosis_H37Rv IA-WLPTLLG-----LGGADLVVVPELAYPTYDVGARLAGTRVLR---AD
MLBr_01488|M.leprae_Br4923 IA-WLPTLLG-----LRAGDVVVVPELAYPTYNVGARLAGARVLR---AD
Mflv_1181|M.gilvum_PYR-GCK VE-LLARTLG--GPVAVEAYGLFVFRDAIAACGVQTVPIGVDDDG---AV
Mvan_5625|M.vanbaalenii_PYR-1 VE-LLTRALG--GPIAVESYGLFVFRDAIAAAGASTVPIGVDDRG---AV
MSMEG_6371|M.smegmatis_MC2_155 VE-LLGKVFGTQRSIAVEAYGLFIFRDALAAMGIATAPIGLDEHG---AV
MAB_0196c|M.abscessus_ATCC_199 VE-ILTRMYGLARPIAVEAYGLFIFRDCIEATGGSSTPIGFDGEG---AV
MMAR_2376|M.marinum_M IERALSANLRPGDRVAVEDPGWANLLDLLAALGLPVEPMGVDDDGPLVAD
MUL_1552|M.ulcerans_Agy99 IERALSANLRPGDRVAVEDPGWANLLDLLAALGLPVEPMGVDDDGPLVAD
MAV_3214|M.avium_104 IERALTAHLRPGDRVAVEDPGWANLLDLLAALGFSAEPVRVDDAGPLPED
TH_4188|M.thermoresistible__bu VDVDAASRLR----LDDDVVHTVVFRRLHAGAPFVSTVVHLPDRA-----
:
Mb1211|M.bovis_AF2122/97 ALTQLGPQSPALLYLNSPSNPTGRVLGVDHLRKVVEWARG--RGVLVVSD
Rv1178|M.tuberculosis_H37Rv ALTQLGPQSPALLYLNSPSNPTGRVLGVDHLRKVVEWARG--RGVLVVSD
MLBr_01488|M.leprae_Br4923 SLTQLGLQSPALFYLNSPSNPTGQVLAVNHLRKMVGWARN--RGVVVVSD
Mflv_1181|M.gilvum_PYR-GCK IADLDDTSARSVLLTPAHHNPLGMPLHPSRRAAVIEWATR--TGGYILDD
Mvan_5625|M.vanbaalenii_PYR-1 VDELDGLDVPAVLLTPAHHNPLGMPLHSSRRTAVIEWAQR--TGGFVLDD
MSMEG_6371|M.smegmatis_MC2_155 ISDLDMLDVPAVLLTPAHHSPHGMPLHPSRRSEVIEWARR--TGGYVIDD
MAB_0196c|M.abscessus_ATCC_199 VSDLENLDTSAALITPAHHFPHGLPLHPARRTAVVDWARR--TDSYVLED
MMAR_2376|M.marinum_M LQRALGRGVRAVVVTSRAQNPTGAALSAARAEELREVLSQGAEEVLLVED
MUL_1552|M.ulcerans_Agy99 LQRALGRGVRAVVVTSRAQNPTGAALSAARAEELREVLSQGAEEVLLVED
MAV_3214|M.avium_104 LARALGRGARALVLTTRAQNPTGAALSAERAAALRDLLSGRADDVLLVED
TH_4188|M.thermoresistible__bu ---AHHLHGCAELRTGAVSTHTVIGLLEPHLEEPIDQAAQ----------
: . * :
Mb1211|M.bovis_AF2122/97 ECYLGLGWDAEPVSVLHPSVCDGDHTGLLAVHSLSKSSSLAGYRAGFVVG
Rv1178|M.tuberculosis_H37Rv ECYLGLGWDAEPVSVLHPSVCDGDHTGLLAVHSLSKSSSLAGYRAGFVVG
MLBr_01488|M.leprae_Br4923 ECYLGLGWDAEPFSVLHPLVCDGNHTGLLAVHSLSKSSSLAGYRAGFVAG
Mflv_1181|M.gilvum_PYR-GCK DYDGEFRYDRQPVGALQALRPD--RVAYLGSTSKSLSQALRLGWMALPAE
Mvan_5625|M.vanbaalenii_PYR-1 DYDGEFRYDRQPVGALQSLRPD--RVAYLGSTSKSLSQTLRLGWMALPDN
MSMEG_6371|M.smegmatis_MC2_155 DYDGEFRYDRQPVGALQSLDPE--HVAYLGSASKSLAQTLRVGWMALPQA
MAB_0196c|M.abscessus_ATCC_199 DYDGEFRYDRQPVGALQSLDPD--RVVYLGSASKSLAPALRLGWMVLPPE
MMAR_2376|M.marinum_M DHCAGIAG-----VPLHTLAGATQHWAFVRSAAKAYGPDLRLAVLAGDQR
MUL_1552|M.ulcerans_Agy99 DHCAGIAG-----VPLHTLAGATQHWAFVRSAAKAYGPDLRLAVLAGDQR
MAV_3214|M.avium_104 DHCAGISG-----AALHPVAGCTTHWAFVRSASKAYGPDLRVAVLAGDQR
TH_4188|M.thermoresistible__bu ----------------------------------------SITVAPADDD
Mb1211|M.bovis_AF2122/97 DLEIVAELLAVRKHAGMMVPAPVQAAMVAALDDDAHERQQRERYAQRRAA
Rv1178|M.tuberculosis_H37Rv DLEIVAELLAVRKHAGMMVPAPVQAAMVAALDDDAHERQQRERYAQRRAA
MLBr_01488|M.leprae_Br4923 DPDLVAELLAVRKHAGMMVPTPVQAAMVAALGDDAHEREQRDRYARRRDV
Mflv_1181|M.gilvum_PYR-GCK LVDQVVETAGGQQFHVDVVSQLTLADFLTAGHYDRHIRRMRMRYRRRRDM
Mvan_5625|M.vanbaalenii_PYR-1 LIDAVLAAAGGQQFYVDAINQLTLADFIAGGHYDRHIRRMRMRYRRRRDV
MSMEG_6371|M.smegmatis_MC2_155 LVEPVIAAAGGQQFNVDAITALTLADFIATGGYDRHIRRMRNRYRRRRDH
MAB_0196c|M.abscessus_ATCC_199 QMDAAVAATGGAQWYVGAISQLTMADFIAGGHYDRHIRRMRGRYRRRRDL
MMAR_2376|M.marinum_M TVERVHGRLRLGPGWVSHILQDLAVRLFADDAATGLVRQAQERYADARGR
MUL_1552|M.ulcerans_Agy99 TVERVHGRLRLGPGWVSHILQDLAVRLFADDAATGLVRQAQERYADARGR
MAV_3214|M.avium_104 TVDRVHGRLRLGPGWVSHVLQQLAVELWSDTAASELVATAERRYADRRRR
TH_4188|M.thermoresistible__bu TADAVGVEPGHPMLRIDRLYSDRTGRPVELSVSHFLPEQYTYRVTLRRE-
: . : * *
Mb1211|M.bovis_AF2122/97 LLPALGSAGFAVDYSDAGLYLWATRGE------PCRDSVAWLAQRGILVA
Rv1178|M.tuberculosis_H37Rv LLPALGSAGFAVDYSDAGLYLWATRGE------PCRDSAAWLAQRGILVA
MLBr_01488|M.leprae_Br4923 LLPAVVSAGFTVDHSEAGLYLWASRGE------PCRDSVEWFAQRGILVA
Mflv_1181|M.gilvum_PYR-GCK LVSALSPFAVGIGGLAAGVNLLVTLPDGAEPELIRRAGEAGIALVGLSMM
Mvan_5625|M.vanbaalenii_PYR-1 LVSALSPFDVGIGGLAAGVNLLVTLPDGAEPEVMRRAGESGLAVSGLSIM
MSMEG_6371|M.smegmatis_MC2_155 LVEALSGFDVGIRGLAAGVNMLLTLPDGAEPEVLRRAGEAGIALSGLALM
MAB_0196c|M.abscessus_ATCC_199 LVQRLAGYNVGVRGLSAGLHLLLTLPEGTESDVLYRAGEAGVALAGLARL
MMAR_2376|M.marinum_M LCAALSARGVAAHG-RSGLNVWIPVPD---------ETAAITRLLNRGWA
MUL_1552|M.ulcerans_Agy99 LCAALSARGVAAHG-RSGLNVWIPVPD---------ETAAITRLLNRGWA
MAV_3214|M.avium_104 LCGALAERGVDAHG-RSGLNVWVRVPD---------EAVAVSRLLGAGWA
TH_4188|M.thermoresistible__bu --------------------------------------------------
Mb1211|M.bovis_AF2122/97 PGDFYGPGGAQHVRVAL---TATDERVAAAVGRLTC----------
Rv1178|M.tuberculosis_H37Rv PGDFYGPGGAQHVRVAL---TATDERVAAAVGRLTC----------
MLBr_01488|M.leprae_Br4923 LGEFYGPGGCQHVRVAL---TASDERIAAVVARLS-----------
Mflv_1181|M.gilvum_PYR-GCK RHPRAGSGVAERDGLIVGFAAAPEHAFPAAVSALCEVLRASGL---
Mvan_5625|M.vanbaalenii_PYR-1 RHPMAGP--ADVDGLIVGFAAPPEHAFSAAVAALCDVLRASGL---
MSMEG_6371|M.smegmatis_MC2_155 RHPLAHPDIPAPDGIIVGFGAPAEHAFRPAVEALRGVLEDVLSP--
MAB_0196c|M.abscessus_ATCC_199 RHPLAGPHIPNPDGVVVSFGTPADHGFGPAVDALCRVLAATGL---
MMAR_2376|M.marinum_M AAPGARFRIRSAPGIRVTIAGLSAEEIEPLADAIAEAIQRTGRPTV
MUL_1552|M.ulcerans_Agy99 AAPGARFRVRSAPGIRVTIAGLSAEEIEPLADAIAGAIQRTGRPTV
MAV_3214|M.avium_104 AAPGARFRMQTPAGIRITIADLTPDEIEPLADAVAQAVRPSGRPIV
TH_4188|M.thermoresistible__bu ----------------------------------------------