For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MADAPASRRPSADDLLVLLAVGRTGRYTTAADELGINHTTISRRIAALEEALGGRVLTRGAGGWELTDLG RDALAAAETVEAAVTSLAADRRRQLEGVVRISATDGFSAYIAAPAAAQVQREHPRIAVELVATTRRASQQ RTSLDLEIVVGAPQVRRAEAIRLGDYCLGLYGARDYLAEHGTPESVADLERYPLVYFIDSMLQVDDLDLA PAFAPAMHESVTSTNVFVHVEATRASAGIGLLPCFMADRHDDLVRVLPETVSVRLSYWLVARAETLRRPV VAVVVDAIRDRMADQRDVLLGVR
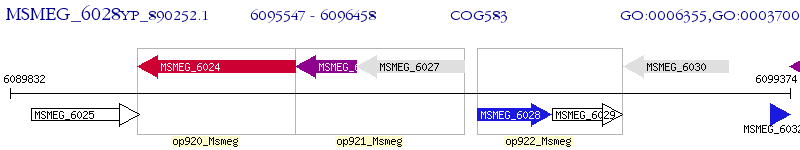
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6028 | - | - | 100% (303) | transcriptional regulator, LysR family protein |
| M. smegmatis MC2 155 | MSMEG_1391 | - | 9e-46 | 35.31% (286) | transcriptional regulator, LysR family protein |
| M. smegmatis MC2 155 | MSMEG_1985 | - | 4e-12 | 27.03% (185) | transcriptional regulator, LysR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2303c | - | 9e-07 | 30.00% (100) | LysR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1470 | - | 1e-132 | 78.67% (300) | LysR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv2282c | - | 9e-07 | 30.00% (100) | LysR family transcriptional regulator |
| M. leprae Br4923 | MLBr_02041 | oxyR | 6e-08 | 28.12% (128) | putative LysR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_0055c | - | 5e-12 | 28.74% (247) | DNA-binding transcriptional regulator CynR |
| M. marinum M | MMAR_2756 | oxyR | 2e-09 | 26.61% (248) | hydrogen peroxide-inducible genes activator, OxyR |
| M. avium 104 | MAV_4979 | - | 4e-07 | 27.73% (119) | transcriptional regulator, LysR family protein, putative |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2998 | oxyR | 8e-11 | 27.02% (248) | hydrogen peroxide-inducible genes activator, OxyR |
| M. vanbaalenii PYR-1 | Mvan_5301 | - | 1e-134 | 81.42% (296) | LysR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2756|M.marinum_M ------------MLATILIGMTDKSYQPTLAGLRAFVAVAEKQHFGSAAN
MUL_2998|M.ulcerans_Agy99 -----------------MIGITDKSYQPTLAGLRAFVAVAEKQHFGSAAN
MLBr_02041|M.leprae_Br4923 --------------------MSDKSYQPTIAGLRAFVAVVEKGHFSAAAS
Mb2303c|M.bovis_AF2122/97 --------------------MPLSSRMPGLTCFEIFLAIAEAGSLGGAAR
Rv2282c|M.tuberculosis_H37Rv --------------------MPLSSRMPGLTCFEIFLAIAEAGSLGGAAR
Mflv_1470|M.gilvum_PYR-GCK MQLPPPAMIKTAGWFCENAEMSPLGRRPSADDLLVLLAVGRTGKYTSAAD
Mvan_5301|M.vanbaalenii_PYR-1 MQLPRAAMAKTAPGICKNAEMSSPVRRPSADDLLVLLAVGRTGRYTTAAD
MSMEG_6028|M.smegmatis_MC2_155 ---------------MADAPAS---RRPSADDLLVLLAVGRTGRYTTAAD
MAB_0055c|M.abscessus_ATCC_199 ---------------------------MELRHIRYLLAVADHANFTRAAE
MAV_4979|M.avium_104 ----------MLLAVSAVGGPESRCVMATLLQLKAFLAVVDQGGFTAASR
: ::*: *:
MMAR_2756|M.marinum_M ALGVSQSTLSQALAALEAGLDLHLVERSTRRVFLTTEGRSLLPHAQAAVE
MUL_2998|M.ulcerans_Agy99 ALGVSQSTLSQALAALEAGLDLHLVERSTRRVFLTTEGRSLLPHAQAAVE
MLBr_02041|M.leprae_Br4923 FLGVRQSTLSQALAALESGLGVQLIERSTRRVFVTTQGRHLLPRAQAAIE
Mb2303c|M.bovis_AF2122/97 ELGLTQQAVSRRLASMEAQIGVRLAIRTTRGSQLTPAGIVVAEWAARLLE
Rv2282c|M.tuberculosis_H37Rv ELGLTQQAVSRRLASMEAQIGVRLAIRTTRGSQLTPAGIVVAEWAARLLE
Mflv_1470|M.gilvum_PYR-GCK ELGLNHTTISRRIAALEQAMGGRVLARVAGGWELTDLGREALAAAEVVET
Mvan_5301|M.vanbaalenii_PYR-1 ELGLNHTTISRRIAALERAMGGRVLARVAGGWELTDLGREALAAAEAVET
MSMEG_6028|M.smegmatis_MC2_155 ELGINHTTISRRIAALEEALGGRVLTRGAGGWELTDLGRDALAAAETVEA
MAB_0055c|M.abscessus_ATCC_199 SLHISQPTLSQQIKQLERTLGVILLDRSGRAVRLTDAGEAYAQHARLALR
MAV_4979|M.avium_104 SLGMSQPAVSRAVAGLERELGSPLLIRHRDRILLNAPGKRALAHARSAVR
* : : ::*: : :* :. : * :. * *
MMAR_2756|M.marinum_M AVEAFTTAAAGEADPLNGSIRFGIIPTVAPYVLPTVLAGL-----TRRLP
MUL_2998|M.ulcerans_Agy99 AVEAFTTAAAGEADPLNGSIRFGIIPTVAPYVLPTVLAGL-----TRRLP
MLBr_02041|M.leprae_Br4923 AMDAFTAAAAGESDPLRGGMRLGLIPTVAPYVLPTMLTGL-----THRLP
Mb2303c|M.bovis_AF2122/97 VADEIDAGLGSLRTEGRQRIRVVASQTIAEQLMPHWMLSLRAADMRRGGT
Rv2282c|M.tuberculosis_H37Rv VADEIDAGLGSLRTEGRQRIRVVASQTIAEQLMPHWMLSLRAADMRRGGT
Mflv_1470|M.gilvum_PYR-GCK AVRSLATDPAG-QRALEGVVRISATDGFSAYIAAPAAAAV-----QRRHP
Mvan_5301|M.vanbaalenii_PYR-1 AVRSLATDAAG-QRALEGVVRISATDGFSAYIAAPAAAAV-----QRRHP
MSMEG_6028|M.smegmatis_MC2_155 AVTSLAADRR---RQLEGVVRISATDGFSAYIAAPAAAQV-----QREHP
MAB_0055c|M.abscessus_ATCC_199 DLDAGERAVHDMHDLSRGHLRIAMTPTFTAYFIGPLVHRF-----HSAHP
MAV_4979|M.avium_104 HLDQMRAEVSA-AAKLRGTLRIASLPYTTELLIAPQLQKF-----SERHP
. :*. : . . .
MMAR_2756|M.marinum_M TLTVRVIEDQTERLLTALREGSLDTALIALPADYGGLTEIPIYEEDFVLA
MUL_2998|M.ulcerans_Agy99 TLTVRVIEDQTERLLTALREGSLDTALIALPADYGGLTEIPIYEEDFVLA
MLBr_02041|M.leprae_Br4923 TLTLRVVEDQTEHLLTALREGALDAAMIALPVETAGIAEIPIYDEDFVLA
Mb2303c|M.bovis_AF2122/97 VPEVILTATNSEHAIAAVRDGIADLGFIENPCPPTGLGSVVVARDELVVV
Rv2282c|M.tuberculosis_H37Rv VPEVILTATNSEHAIAAVRDGIADLGFIENPCPPTGLGSVVVARDELVVV
Mflv_1470|M.gilvum_PYR-GCK GVAVEIVATTRR---ATQQRSGMDVEVVVGEPKVRRARALRLSDYCLGLY
Mvan_5301|M.vanbaalenii_PYR-1 KVAVEIVATTRR---ATQQRSGLDIEVVVGEPKVHRAKALRLGDYCLGLY
MSMEG_6028|M.smegmatis_MC2_155 RIAVELVATTRR---ASQQRTSLDLEIVVGAPQVRRAEAIRLGDYCLGLY
MAB_0055c|M.abscessus_ATCC_199 GITLTVRETTQDRIEADLLADRLDLGIAFAGPHPAGIDHCALFTETLRLV
MAV_4979|M.avium_104 SVEIRVLEGGEPEIRDWLDLGAADAGVISLP--ATGLSAAFLAAQDMVAV
: : * . : :
MMAR_2756|M.marinum_M LPPGHPMADR-HGVPVAALSELPLLLLDEGHCLRDQALDVCQN---AGVR
MUL_2998|M.ulcerans_Agy99 LPPGHPMADR-HGVPVAALSELPLLLLDEGHCLRDQALDVCQN---AGVR
MLBr_02041|M.leprae_Br4923 LPPGHPLSGK-RRVPTTALAQLPLLLLDEGHCLRDQTLDICRK---SGVQ
Mb2303c|M.bovis_AF2122/97 VPPGHKWARRSRVVSARELAQTPLVTREPNSGIRDSLTAALRDTLGEDMQ
Rv2282c|M.tuberculosis_H37Rv VPPGHKWARRSRVVSARELAQTPLVTREPNSGIRDSLTAALRDTLGEDMQ
Mflv_1470|M.gilvum_PYR-GCK GSR-DYLATHGTPATIAELQNHPLVYFIDSMLQVDDLDLATSF-----TP
Mvan_5301|M.vanbaalenii_PYR-1 ASR-DYLAEHGHPTSVADLQRFPLVYFIDAMLQVDDLDLATSF-----AP
MSMEG_6028|M.smegmatis_MC2_155 GAR-DYLAEHGTPESVADLERYPLVYFIDSMLQVDDLDLAPAF-----AP
MAB_0055c|M.abscessus_ATCC_199 VGTGHRLAMRHRSLPVAQLSREALGLLSPDFATRQDIDAYFAG----LGV
MAV_4979|M.avium_104 LPSEHRLAAF-SEVHYADLAKEPFIRSTGGCAHVFMAVADQVG------V
. : * . .:
MMAR_2756|M.marinum_M AELADTRAASLATAVQCVNGGLGVTLIPQSAVSVEAARSRVGLAHFAAPT
MUL_2998|M.ulcerans_Agy99 AEHADTRADSLATAVQCVNGGLGVTLIPQSAVSVEAARSRVGLTHFAAPT
MLBr_02041|M.leprae_Br4923 AELANTRAASLATAVQCVTGGLGVTLLPQSAAPVESVRSKLGLAQFAAPC
Mb2303c|M.bovis_AF2122/97 QAPPVLELSSAAAVRAAVLAGAGPAAMSRLAIADDLAFGRLLAVDIPALN
Rv2282c|M.tuberculosis_H37Rv QAPPVLELSSAAAVRAAVLAGAGPAAMSRLAIADDLAFGRLLAVDIPALN
Mflv_1470|M.gilvum_PYR-GCK AMRESVTSTNVFVHVEATRAAAGIGLLPCFMADRHDDLVRVLPGQVSLTL
Mvan_5301|M.vanbaalenii_PYR-1 AMRESVTSTNVFVHVEATRAAAGIGLLPCFMADRHPDLERVLAAAVSIRL
MSMEG_6028|M.smegmatis_MC2_155 AMHESVTSTNVFVHVEATRASAGIGLLPCFMADRHDDLVRVLPETVSVRL
MAB_0055c|M.abscessus_ATCC_199 APHIAVEANSISALLEFVKRGSLATVLPDAIAHEHSDLH---LVPLSPPL
MAV_4979|M.avium_104 EFDVAFEAHGLMAVLEIVTAGLGVSIVPVAAVARP-----RAGIAVKRLV
. . . . :. .
MMAR_2756|M.marinum_M PGRRIGLVYRASSGRDEAYRQLAAIMGESISNEQQVRLVN-
MUL_2998|M.ulcerans_Agy99 PGRRIGLVYRASSGRDEAYRQLAAIMGESISNEQQVRLVN-
MLBr_02041|M.leprae_Br4923 PGRRIGLAFRSASGRSASYRQIAKIIGELISTEHHVRLVA-
Mb2303c|M.bovis_AF2122/97 LRRQLRAIWVG--GRTPPAGAIRDLLSHITSRST-------
Rv2282c|M.tuberculosis_H37Rv LRRQLRAIWVG--GRTPPAGAIRDLLSHITSRST-------
Mflv_1470|M.gilvum_PYR-GCK T---YWLVTRAETLRRPEVAAVVDAIRDRVRDRHHMLLGAG
Mvan_5301|M.vanbaalenii_PYR-1 T---YWLVTRAETLRRPEVAAVVDAIRDRVREQHDVLLGAG
MSMEG_6028|M.smegmatis_MC2_155 S---YWLVARAETLRRPVVAVVVDAIRDRMADQRDVLLGVR
MAB_0055c|M.abscessus_ATCC_199 PIRTVELLRRSSAYHSAAARAFTEMVHAGARGR--------
MAV_4979|M.avium_104 PKTTRNLAVAVSASAGSAARAFLEQIAALDGSERAGA----
. :