For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MADVGDLEFFVTLAAAGTMTEAARHWGVSVSVVSRRLKALEERLGTPLVHRRARGLELTAEGQQYHVRGS EILQQLKDLESTLNPDPKDLTGSIRVISTVGLGRVHIAPLLHDFRRRHRSVEISLELTSLPLSASVPGFD IAIQVGRVRDSSLAIRQLLPNRRVVVASPDYLDAHGAPADLDDLKNHELLVVRENEGESIWRFVKDGRET AIPVRGGLICNDGIAVTEWCLAGAGLAMRSIWHVAPYVREGRLVHVLPDVETPEADVVALFDAGVRTSPR VRTLLSFLRKQLAQRCIPLPTRQAGAALHW
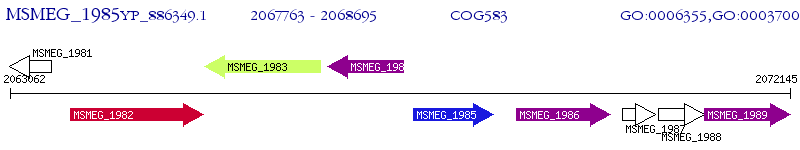
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1985 | - | - | 100% (310) | transcriptional regulator, LysR family protein |
| M. smegmatis MC2 155 | MSMEG_1391 | - | 3e-19 | 28.52% (298) | transcriptional regulator, LysR family protein |
| M. smegmatis MC2 155 | MSMEG_6849 | - | 8e-13 | 27.84% (194) | LysR-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0384 | - | 3e-09 | 27.67% (318) | LysR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1470 | - | 2e-15 | 28.11% (185) | LysR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0377 | - | 2e-08 | 28.01% (307) | LysR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2680c | - | 1e-10 | 35.54% (121) | LysR family transcriptional regulator |
| M. marinum M | MMAR_0653 | - | 2e-08 | 27.67% (318) | LysR family transcriptional regulator |
| M. avium 104 | MAV_1538 | - | 3e-11 | 25.71% (280) | transcriptional regulator, LysR family protein |
| M. thermoresistible (build 8) | TH_0172 | - | 9e-05 | 29.46% (129) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY |
| M. ulcerans Agy99 | MUL_2698 | - | 7e-08 | 23.88% (268) | LysR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5301 | - | 4e-13 | 28.11% (185) | LysR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0384|M.bovis_AF2122/97 ---------------------------MTPAQLRAYSAVVRLGSVRAAAA
Rv0377|M.tuberculosis_H37Rv ---------------------------MTPAQLRAYSAVVRLGSVRAAAA
MMAR_0653|M.marinum_M ---------------------------MTPAQLRAYSAVVRLGSVRAAAT
TH_0172|M.thermoresistible__bu ---------------------------MTPAQLRAFSAVVRLGSVRAAAQ
Mflv_1470|M.gilvum_PYR-GCK MQLPPPAMIKTAGWFCENAEMSPLGRRPSADDLLVLLAVGRTGKYTSAAD
Mvan_5301|M.vanbaalenii_PYR-1 MQLPRAAMAKTAPGICKNAEMSSPVRRPSADDLLVLLAVGRTGRYTTAAD
MUL_2698|M.ulcerans_Agy99 --------------------MPLSPRMPELASFEIFLAIAQTGSLGAAAR
MAV_1538|M.avium_104 ---------------------------MAVGDYEWFITLAELQHVTAAAE
MAB_2680c|M.abscessus_ATCC_199 --------------------MDQWTSQLAPQLRALVELAAHDGHMTQAAL
MSMEG_1985|M.smegmatis_MC2_155 --------------------------MADVGDLEFFVTLAAAGTMTEAAR
**
Mb0384|M.bovis_AF2122/97 ELGLSDAGVSMHVAALRKELDDPLFTRTGAGLAFTPGGLRLASRAVEILG
Rv0377|M.tuberculosis_H37Rv ELGLSDAGVSMHVAALRKELDDPLFTRTGAGLAFTPGGLRLASRAVEILG
MMAR_0653|M.marinum_M ELGLSDAGVSMHVAALRKELDDPLFTRTGAGLAFTPGGLRLASRAVEILG
TH_0172|M.thermoresistible__bu ELGMSDAGISMHIAQLRKELDDPLFHRTKNGLSFTPGGLRLASRAVDILG
Mflv_1470|M.gilvum_PYR-GCK ELGLNHTTISRRIAALEQAMGGRVLARVAGGWELTDLGREALAAAEVVET
Mvan_5301|M.vanbaalenii_PYR-1 ELGLNHTTISRRIAALERAMGGRVLARVAGGWELTDLGREALAAAEAVET
MUL_2698|M.ulcerans_Agy99 ELGLTQQAVSKRLAAMEAHTGVTLAVRTTRGSQLTPAGLIVSEWAARLLE
MAV_1538|M.avium_104 QLHVAQPTLTRMLARLEQRLGVALFDRHGRRLSLNTYGRIFYEHARRAQL
MAB_2680c|M.abscessus_ATCC_199 TLGIPQSSMSRRIHALQAALGVPLLIHDGRTVRLTPEARLLAARTREPLA
MSMEG_1985|M.smegmatis_MC2_155 HWGVSVSVVSRRLKALEERLGTPLVHRRARGLELTAEGQQYHVRGSEILQ
: :: : :. . : : :. .
Mb0384|M.bovis_AF2122/97 LQQQTAIEVTEAA-HGRRLLRIAASSAFAEHAAPGLIELFS---SRADDL
Rv0377|M.tuberculosis_H37Rv LQQQTAIEVTEAA-HGRRLLRIAASSAFAEHAAPGLIELFS---SRADDL
MMAR_0653|M.marinum_M LQQQTAIEVTEAA-HGRRLLRIAASSTFAEHAAPGLIELFS---SRADDL
TH_0172|M.thermoresistible__bu LQRQTAIEVTEAA-HGRRLLRIAASHAFAEHAAPGLIDLFS---SRADDL
Mflv_1470|M.gilvum_PYR-GCK AVRSLATDPAGQR-ALEGVVRISATDGFSAYIAAPAAAAVQ---RRHPGV
Mvan_5301|M.vanbaalenii_PYR-1 AVRSLATDAAGQR-ALEGVVRISATDGFSAYIAAPAAAAVQ---RRHPKV
MUL_2698|M.ulcerans_Agy99 VAQEIDAGLGSLRKEGRERIRVAASQTIAEQLMPHWLLALQDAAKRRGTT
MAV_1538|M.avium_104 ELDSARRAIADLADPAVGEIRLAYLSSFGSTVVPRLIARFK-----ESSP
MAB_2680c|M.abscessus_ATCC_199 ELDHTLSEVTGDADPERGTVRFGFPLTMGSGHIPDLLAAFR-----HRHP
MSMEG_1985|M.smegmatis_MC2_155 QLKDLESTLNPDPKDLTGSIRVISTVGLGRVHIAPLLHDFR---RRHRSV
:*. :. . .
Mb0384|M.bovis_AF2122/97 SVELSVHPTSRFRELICSRAVDIAIG-PASESSIGSDGSIFLRPFLKYQI
Rv0377|M.tuberculosis_H37Rv SVELSVHPTSRFRELICSRAVDIAIG-PASESSIGSDGSIFLRPFLKYQI
MMAR_0653|M.marinum_M SVELSVHPTSQFRDLICSRAVDIAIG-PASESSIGSDTSLFVRPFLKYQI
TH_0172|M.thermoresistible__bu SVELTVEPVSRFRDLLESRAVDVTLG-PAVRE---PGPAIVVRPFLNYQI
Mflv_1470|M.gilvum_PYR-GCK AVEIVAT-TRRATQQRSGMDVEVVVGEPKVRRARALRLSDYCLGLYGSRD
Mvan_5301|M.vanbaalenii_PYR-1 AVEIVAT-TRRATQQRSGLDIEVVVGEPKVHRAKALRLGDYCLGLYASRD
MUL_2698|M.ulcerans_Agy99 APQVILTATNSDHAIAAVRDGTADLG-FVENPGPPTGLGSRVVGHDDLVV
MAV_1538|M.avium_104 RVTFTLEEGAAESIADRVRSGGVDIG--VVSPRPVERTLAWRSLFRQRLG
MAB_2680c|M.abscessus_ATCC_199 GIRVLLKQAHGSELGAQLLTGDLDLA----VVIPAPERLHHITIGAQHIH
MSMEG_1985|M.smegmatis_MC2_155 EISLELTSLPLSASVPG-FDIAIQVG-----RVRDSSLAIRQLLPNRRVV
. :.
Mb0384|M.bovis_AF2122/97 ITVVAPNSPLAAGIPMPALLRHQQWMLGPSAGSVDGEIATMLRGLA----
Rv0377|M.tuberculosis_H37Rv ITVVAPNSPLAAGIPMPALLRHQQWMLGPSAGSVDGEIATMLRGLA----
MMAR_0653|M.marinum_M ITVVAPNSPLAVGIPTPSLLRQQQWMLGPPAGSVAGEIATMLRDLA----
TH_0172|M.thermoresistible__bu IAVTSPSNPLAAATPTIAQLRAQSWMLGPSAGGIDGDVAAMLRRLE----
Mflv_1470|M.gilvum_PYR-GCK YLATHGTPATIAELQNHPLVYFIDSMLQVDDLDLATSFTPAMRESV----
Mvan_5301|M.vanbaalenii_PYR-1 YLAEHGHPTSVADLQRFPLVYFIDAMLQVDDLDLATSFAPAMRESV----
MUL_2698|M.ulcerans_Agy99 VVPPGHKWTRRPSPVTARELAEMPLVTREPRSGIRDSLTVALRQVLGEDM
MAV_1538|M.avium_104 VAVPDGHRFARGAAVSMVDLADEPFVTMHPGFGMRRLLDELCAAAQ----
MAB_2680c|M.abscessus_ATCC_199 IAVPEDHRLAGAAQVRLRELSGETFIANPPSYNLRQLTERWCREAG----
MSMEG_1985|M.smegmatis_MC2_155 VASPDYLDAHGAPADLDDLKNHELLVVRENEGESIWRFVKDGRETA----
:
Mb0384|M.bovis_AF2122/97 IPESQQRIFQSDAAALEEVMRVGGATLAIGFAVAKDLAAGRLVHVTGPGL
Rv0377|M.tuberculosis_H37Rv IPESQQRIFQSDAAALEEVMRVGGATLAIGFAVAKDLAAGRLVHVTGPGL
MMAR_0653|M.marinum_M IPESQQRIFQSDAAALEEVMRVGGATLTIGFAVAKDLAAGRLVHVTSPGL
TH_0172|M.thermoresistible__bu IPEAAQRIFQSDSAAQEEVQRIGGVTLSIGFAVAKDLAAGRLARIKG--V
Mflv_1470|M.gilvum_PYR-GCK TSTN---VFVH----VEATRAAAGIGLLPCFMADR---HDDLVRVLPGQV
Mvan_5301|M.vanbaalenii_PYR-1 TSTN---VFVH----VEATRAAAGIGLLPCFMADR---HPDLERVLAAAV
MUL_2698|M.ulcerans_Agy99 VQASPVLELSSAAAMRAAVLAGAGPAAMSRLAIADDLAVGRLRAISIPGL
MAV_1538|M.avium_104 --FRPRVALQAPNLTTAASLVAAGLGISLVPIDGS----SYPSGVSVKPL
MAB_2680c|M.abscessus_ATCC_199 --YAPNVTIEVTEFSTIRELIGRGLGVALLPHDDR-----ALPGIREIPL
MSMEG_1985|M.smegmatis_MC2_155 IPVRGGLICNDGIAVTEWCLAGAGLAMRSIWHVAPYVREGRLVHVLPDVE
* :
Mb0384|M.bovis_AF2122/97 DRAGEWCVATLAPSARQPAVSELVGFIST-PRCIQAMIRGSGVGVTRFRP
Rv0377|M.tuberculosis_H37Rv DRAGEWCVATLAPSARQPAVSELVGFIST-PRCIQAMIPGSGVGVTRFRP
MMAR_0653|M.marinum_M DKAGEWCVATLPPAARQPAVSELVRFITT-PRCIQAMIRGSGVGVTRFRP
TH_0172|M.thermoresistible__bu QATAEWCVSTLSSAARQPAASELVRFITT-PRCTQAMIRGTSEGVTRFRP
Mflv_1470|M.gilvum_PYR-GCK SLTLTYWLVTRAETLRRPEVAAVVDAIRDRVRDRHHMLLGAG--------
Mvan_5301|M.vanbaalenii_PYR-1 SIRLTYWLVTRAETLRRPEVAAVVDAIRDRVREQHDVLLGAG--------
MUL_2698|M.ulcerans_Agy99 DLRRQLRAIWVGG--RVPPAGAIRELLSH--------ITSGGE-------
MAV_1538|M.avium_104 ADADAYRDVGMIWDSGRPLPRSARDFIAG-----AAAVGHGGW-------
MAB_2680c|M.abscessus_ATCC_199 AGDGYHRAIALAWGTATRAAPTRRLNDFLLQHFSQAEVNSAAIRKG----
MSMEG_1985|M.smegmatis_MC2_155 TPEADVVALFDAGVRTSPRVRTLLSFLRKQLAQRCIPLPTRQAGAALHW-
:
Mb0384|M.bovis_AF2122/97 KVHVTLWS
Rv0377|M.tuberculosis_H37Rv KVHVTLWS
MMAR_0653|M.marinum_M KVHVTLWS
TH_0172|M.thermoresistible__bu KVHVTLWS
Mflv_1470|M.gilvum_PYR-GCK --------
Mvan_5301|M.vanbaalenii_PYR-1 --------
MUL_2698|M.ulcerans_Agy99 --------
MAV_1538|M.avium_104 --------
MAB_2680c|M.abscessus_ATCC_199 --------
MSMEG_1985|M.smegmatis_MC2_155 --------