For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDFHGKTALVTGASAGLGAAVAKLLAARGASVFGIARDADRMAEVFTEVPGGRYASVDISSSQGCDQAV EQCVEQFGQLDTLINVAGFHQMRHTVSVTDEDWAKDLAVNLNGPFFLSRAALPHLLQAGGNIVNVASIAG VEGEVYSAGYCAAKHGLVGLTRALAVEFTKERLRVNAVCPGGMLTAQTTEFSAPDNADWDLIMRIAAPRG FMDVADVAKTIAFLASDDAAAVHGAVYIVDAGKTAG
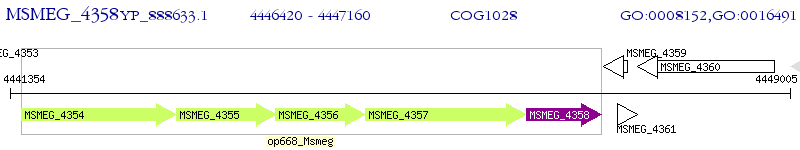
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4358 | - | - | 100% (246) | D-beta-hydroxybutyrate dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3607 | - | 7e-33 | 33.20% (244) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3994 | - | 2e-30 | 33.99% (253) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2025 | fabG3 | 3e-29 | 33.33% (243) | 20-beta-hydroxysteroid dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2757 | - | 1e-111 | 78.46% (246) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2002 | fabG3 | 3e-29 | 33.33% (243) | 20-beta-hydroxysteroid dehydrogenase |
| M. leprae Br4923 | MLBr_02565 | fabG | 2e-24 | 33.90% (236) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. abscessus ATCC 19977 | MAB_0073c | - | 1e-28 | 30.92% (262) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_0302 | - | 1e-31 | 35.42% (240) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_0087 | - | 5e-95 | 70.33% (246) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. thermoresistible (build 8) | TH_3962 | - | 5e-29 | 32.85% (274) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_4820 | - | 8e-30 | 35.00% (240) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_3777 | - | 1e-110 | 78.05% (246) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2757|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3777|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4358|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0087|M.avium_104 --------------------------------------------------
MMAR_0302|M.marinum_M --------------------------------------------------
MUL_4820|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 MAPKCSSDLFSQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLI
Mb2025|M.bovis_AF2122/97 --------------------------------------------------
Rv2002|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0073c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3962|M.thermoresistible__bu --------------------------------------------------
Mflv_2757|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3777|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4358|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0087|M.avium_104 --------------------------------------------------
MMAR_0302|M.marinum_M --------------------------------------------------
MUL_4820|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GGDGRLVEPLRATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELK
Mb2025|M.bovis_AF2122/97 --------------------------------------------------
Rv2002|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0073c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3962|M.thermoresistible__bu --------------------------------------------------
Mflv_2757|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3777|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4358|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0087|M.avium_104 --------------------------------------------------
MMAR_0302|M.marinum_M --------------------------------------------------
MUL_4820|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GLHNFFTPLLRNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGK
Mb2025|M.bovis_AF2122/97 --------------------------------------------------
Rv2002|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0073c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3962|M.thermoresistible__bu --------------------------------------------------
Mflv_2757|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3777|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4358|M.smegmatis_MC2_155 --------------------------------------------------
MAV_0087|M.avium_104 --------------------------------------------------
MMAR_0302|M.marinum_M --------------------------------------------------
MUL_4820|M.ulcerans_Agy99 --------------------------------------------------
MLBr_02565|M.leprae_Br4923 ELRRGATVALVYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAAD
Mb2025|M.bovis_AF2122/97 --------------------------------------------------
Rv2002|M.tuberculosis_H37Rv --------------------------------------------------
MAB_0073c|M.abscessus_ATCC_199 --------------------------------------------------
TH_3962|M.thermoresistible__bu --------------------------------------------------
Mflv_2757|M.gilvum_PYR-GCK -------MGQLSGKVAMVTGASAGLGAETAKLFAERGATVFGIARDAD--
Mvan_3777|M.vanbaalenii_PYR-1 -------MSQLSGKVALVTGASAGLGAATAKLFAERGATVFGVARDGS--
MSMEG_4358|M.smegmatis_MC2_155 -------MTDFHGKTALVTGASAGLGAAVAKLLAARGASVFGIARDAD--
MAV_0087|M.avium_104 -------MAALQDKVALVTGASSGLGAETAKLFSSRGATVFGIGRDTA--
MMAR_0302|M.marinum_M ------------MQVAIITGATGGIGFGCAAKLAEMGMAVLGTGRDER--
MUL_4820|M.ulcerans_Agy99 ------------MQVAIITAATGGIGFGCAAKFAEMSMAVVGTARDER--
MLBr_02565|M.leprae_Br4923 STPPTNWDRPLENKVAIVTGAARGIGAAIAEVVARDGARVVAIDVES---
Mb2025|M.bovis_AF2122/97 ------MSGRLIGKVALVSGGARGMGASHVRAMVAEGAKVVFGDILD---
Rv2002|M.tuberculosis_H37Rv ------MSGRLIGKVALVSGGARGMGASHVRAMVAEGAKVVFGDILD---
MAB_0073c|M.abscessus_ATCC_199 -------MTRLGGRVAVVTGAGKGMGRTHCERLAEEGSDVIAIDLPG---
TH_3962|M.thermoresistible__bu ----LSIMGAFTGKVAFVTGAARGQGRSHALRLAREGAAIVALDIAADID
:.*.::.. * * . . :.
Mflv_2757|M.gilvum_PYR-GCK -----------RMAEVFADVPG---GSFRSVDVTSADECRRAVAECVAEF
Mvan_3777|M.vanbaalenii_PYR-1 -----------RMAEVFADIPG---GSFSSVDITSPAACAQAVSECVARF
MSMEG_4358|M.smegmatis_MC2_155 -----------RMAEVFTEVPG---GRYASVDISSSQGCDQAVEQCVEQF
MAV_0087|M.avium_104 -----------RLAEVFAGIER---GGFASVDIASPQACTEAVAQCVRDF
MMAR_0302|M.marinum_M -----------RLAELTQLPGGPDRIATHPVDLTDDDAPGQLTRAALERW
MUL_4820|M.ulcerans_Agy99 -----------RLAELTQLPGGPDRIATHPVDPTDDDAPGQLTRAALDRW
MLBr_02565|M.leprae_Br4923 ------------AAEALDETANRVGGTTLSLDVTADDAVDKITEHLHDYQ
Mb2025|M.bovis_AF2122/97 ---------EEGKAVAAELADAAR---YVHLDVTQPAQWTAAVDTAVTAF
Rv2002|M.tuberculosis_H37Rv ---------EEGKAVAAELADAAR---YVHLDVTQPAQWTAAVDTAVTAF
MAB_0073c|M.abscessus_ATCC_199 --------SDDLAQTAAAVEEHGRRCVWAFADVRDAAALTEAIDTGVRML
TH_3962|M.thermoresistible__bu ECFFPLATPEDLQETVRLVEEDGGRILARRGDVRNYDDVKSAVDDGIAQF
*
Mflv_2757|M.gilvum_PYR-GCK G-RLDVLANIAGFHK-MRHTASMTDDDWATDLSVNLNGPFFLCRAALP-H
Mvan_3777|M.vanbaalenii_PYR-1 G-RLDVLANIAGFHK-MRHTVSMTDADWEADLSVNLNGPFYLCRAALP-H
MSMEG_4358|M.smegmatis_MC2_155 G-QLDTLINVAGFHQ-MRHTVSVTDEDWAKDLAVNLNGPFFLSRAALP-H
MAV_0087|M.avium_104 G-GLDVLVNVAGIHR-MRRTESMTDEDWEHDLAVNLNGPFFLCRAALP-Q
MMAR_0302|M.marinum_M G-RIDFLINAAGVGS-PKPVHETDDESLDYVLSLMLRAPFRLTREVLP--
MUL_4820|M.ulcerans_Agy99 G-RIDFLINAAGEGS-PKSVHETDDESLDYVLSLMLRAPFRLTREVLP--
MLBr_02565|M.leprae_Br4923 GGHADILVNNAGITR-DKLLANMDDCRWDAVLEVNLLAPQRLTEGLVGNG
Mb2025|M.bovis_AF2122/97 G-GLHVLVNNAGILN-IGTIEDYALTEWQRILDVNLTGVFLGIRAVVK-P
Rv2002|M.tuberculosis_H37Rv G-GLHVLVNNAGILN-IGTIEDYALTEWQRILDVNLTGVFLGIRAVVK-P
MAB_0073c|M.abscessus_ATCC_199 G-RLDIVVANAGVYDGLAPSCELTEDSWRRALDVNLTGAWHTVKAGAA-H
TH_3962|M.thermoresistible__bu G-PIGIVVANAGIGTAYKKTWEIPEADWRAALDVNLTGVFHTVKAAVP-S
* : ** . * : * . .
Mflv_2757|M.gilvum_PYR-GCK LLETG--GNIVNVASIAGVEGEVYSAGYCAAKHGLVGLTRALAIEFTKEK
Mvan_3777|M.vanbaalenii_PYR-1 LLESG--GNIVNVASIAGVEGEVYSAGYCAAKHGLIGLTRALAVEFTKER
MSMEG_4358|M.smegmatis_MC2_155 LLQAG--GNIVNVASIAGVEGEVYSAGYCAAKHGLVGLTRALAVEFTKER
MAV_0087|M.avium_104 LLERG--GNIVNVSSIAGVEGQAYSAGYCAAKHGLIGLTRALAVEYTADR
MMAR_0302|M.marinum_M HLRPG--SAIINITSTFAVVGGLRGGAYSAAKGGLTALTTHIACQYGAQG
MUL_4820|M.ulcerans_Agy99 HLRPG--SAIINITSTFAVVGGLRGGAYSAAKGGLAALTTHIACQYGAQG
MLBr_02565|M.leprae_Br4923 SISKG--GRVIVLSSMAGIAGNRGQTNYATAKAGVIGLTQALAPGLAEKG
Mb2025|M.bovis_AF2122/97 MKEAG-RGSIINISSIEGLAGTVACHGYTATKFAVRGLTKSTALELGPSG
Rv2002|M.tuberculosis_H37Rv MKEAG-RGSIINISSIEGLAGTVACHGYTATKFAVRGLTKSTALELGPSG
MAB_0073c|M.abscessus_ATCC_199 LLD---GGSVIIISSTNGLKGTANTAHYTASKHAVVGLARTAANELGPQG
TH_3962|M.thermoresistible__bu MIEAGRGGSITLISSSAGLKGYQHLAPYVSSKHAVIGLMRTLANELAEHN
. : ::* .: * * ::* .: .* *
Mflv_2757|M.gilvum_PYR-GCK LRVNAICPGGMPTAQTTEFE------------APENADWDLIMRIASPRG
Mvan_3777|M.vanbaalenii_PYR-1 LRVNAICPGGMPTAQTTEFE------------APENADWDLIMRIASPRG
MSMEG_4358|M.smegmatis_MC2_155 LRVNAVCPGGMLTAQTTEFS------------APDNADWDLIMRIAAPRG
MAV_0087|M.avium_104 LRVNAVCPGGMLTPQIERFS------------APDDPNYDLILRTAAPRG
MMAR_0302|M.marinum_M IRCNAVAPG-VIQTAMTQHR------------LADERFRRINVEMTPHQR
MUL_4820|M.ulcerans_Agy99 IRCNAVAPG-VIQTAMTQHR------------LADERFRRLNVEITPHQR
MLBr_02565|M.leprae_Br4923 ITINAVAPG-FIETQMTAAI------------PLAT--REVGRRLNSLLQ
Mb2025|M.bovis_AF2122/97 IRVNSIHPGLVKTPMTDWVP--------------EDIFQTALGR------
Rv2002|M.tuberculosis_H37Rv IRVNSIHPGLVKTPMTDWVP--------------EDIFQTALGR------
MAB_0073c|M.abscessus_ATCC_199 IRVNTVHPGAVSTGMILNPAVIARLRPDLEHPTVEDAAEALAARTLLPVP
TH_3962|M.thermoresistible__bu IRVNALCPGSVRTPMIINEPTFKVFRPDLDEPTEADAMEVFRQMPALPVD
: *:: ** .
Mflv_2757|M.gilvum_PYR-GCK FMATEDVAKTIAFLASDDAAAVHGAVYRVDNGKGAG--------------
Mvan_3777|M.vanbaalenii_PYR-1 FMATDDVAKTIAFLASDDAAAIHGAVYRVDNGKGAG--------------
MSMEG_4358|M.smegmatis_MC2_155 FMDVADVAKTIAFLASDDAAAVHGAVYIVDAGKTAG--------------
MAV_0087|M.avium_104 MMRPLDVANVIAFLASDAAAAVHGAVYRVDNGKGAG--------------
MMAR_0302|M.marinum_M LGTVDDVANTVAFLCSEGGAFINGQTIVVDGGWSSTKYLSDFALTAQWSP
MUL_4820|M.ulcerans_Agy99 LGTVDDVANTVAFLCSEGGAFINGQTIVVDGG------------------
MLBr_02565|M.leprae_Br4923 GGLPVDVAETIAYFAIPASNAVTGNVIRVCGQAMLGA-------------
Mb2025|M.bovis_AF2122/97 AAEPVEVSNLVVYLASDESSYSTGAEFVVDGGTVAGLAHNDFGAVEVSSQ
Rv2002|M.tuberculosis_H37Rv AAEPVEVSNLVVYLASDESSYSTGAEFVVDGGTVAGLAHNDFGAVEVSSQ
MAB_0073c|M.abscessus_ATCC_199 WVEAVDISNAVVFLASDEARYITGTQLVVDAGLTQKA-------------
TH_3962|M.thermoresistible__bu VLEPEDISEMVAFLASDAARYVTGLAIPVDAGQLVK--------------
:::: :.::. . * *
Mflv_2757|M.gilvum_PYR-GCK ------
Mvan_3777|M.vanbaalenii_PYR-1 ------
MSMEG_4358|M.smegmatis_MC2_155 ------
MAV_0087|M.avium_104 ------
MMAR_0302|M.marinum_M AQDDSK
MUL_4820|M.ulcerans_Agy99 ------
MLBr_02565|M.leprae_Br4923 ------
Mb2025|M.bovis_AF2122/97 PEWVT-
Rv2002|M.tuberculosis_H37Rv PEWVT-
MAB_0073c|M.abscessus_ATCC_199 ------
TH_3962|M.thermoresistible__bu ------