For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAKATSSVLPPAERILAAAADLLRAGGIDAVSTRAVATEAGVQPPTIYRQFGDKDGLLDAVTRHVLESYI DRKRLLTDPDADPALILRELWDLHVEFGMQQPHCYLLTYGQGRRTSAADETVSILGEVIGRLGAAGRLTM SVRRATDYFRASGTGFVMSQLSLASEERDAELSGIIFEATLAAVSTDGRRGRGRKANDVRARAVALREAL PESRTPALSAAEQGLLAEWLDRLADQA
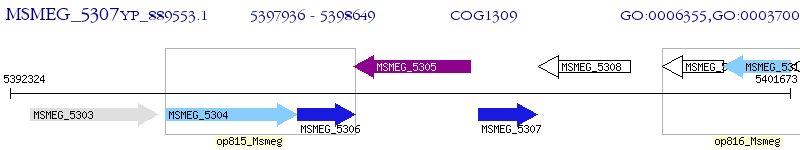
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5307 | - | - | 100% (237) | TetR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_5177 | - | 3e-11 | 38.94% (113) | regulatory protein, TetR |
| M. smegmatis MC2 155 | MSMEG_2557 | - | 1e-06 | 52.08% (48) | transcriptional regulator, TetR family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0672c | - | 2e-07 | 30.61% (98) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0941 | - | 7e-19 | 32.61% (230) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0653c | - | 2e-07 | 30.61% (98) | TetR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2061c | - | 5e-08 | 28.07% (171) | TetR family transcriptional regulator |
| M. marinum M | MMAR_4304 | - | 8e-09 | 37.27% (110) | TetR family transcriptional regulator |
| M. avium 104 | MAV_1669 | - | 4e-07 | 26.02% (196) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_4549 | - | 2e-39 | 43.61% (227) | PUTATIVE putative TetR-family protein transcriptional regulator |
| M. ulcerans Agy99 | MUL_0990 | - | 5e-08 | 36.36% (110) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_4572 | - | 3e-09 | 34.45% (119) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4304|M.marinum_M ---MSMGRLRRARAPRGSGDLLRHEILDAATELLLQTRQARAVSIRSVAE
MUL_0990|M.ulcerans_Agy99 --------MRRARAPRGSGDLLRHEILDAATELLLQTRQARAVSIRSVAE
Mvan_4572|M.vanbaalenii_PYR-1 MADVSAPRQRRHRAPRGAGEQLRAEILDAATDLLLETGDEKAVSIRSVAQ
Mb0672c|M.bovis_AF2122/97 -----------MTSQTG----VRDELLHAGVRLLDDHG-PDALQTRKVAA
Rv0653c|M.tuberculosis_H37Rv -----------MTSQTG----VRDELLHAGVRLLDDHG-PDALQTRKVAA
MSMEG_5307|M.smegmatis_MC2_155 -----------MAKATSSVLPPAERILAAAADLLRAGG-IDAVSTRAVAT
TH_4549|M.thermoresistible__bu -----------VDGLTEQQREQRERILRAAAEILAEQG-RDAVTTRNVSA
MAB_2061c|M.abscessus_ATCC_199 -----------------MAGLRRKLLLEAAADLFARQG-FHAVGIDDIGA
MAV_1669|M.avium_104 -----------------MKSDRRLQLLSAAERLFAERG-FLAVRLEDIGA
Mflv_0941|M.gilvum_PYR-GCK ------------------MDSGRRRLIDAGLALLTEQA-DREPSTRELYE
:: *. :: :
MMAR_4304|M.marinum_M RVGVTPPSIYLHFQDKDALLDAVCARYLARLDEEME--RAAMGHTCVVEV
MUL_0990|M.ulcerans_Agy99 RVGVTSPSIYLHFQDKDALLDAVCARYLARLDEEME--RAAMGHTCVVEV
Mvan_4572|M.vanbaalenii_PYR-1 RVGVTPPSIYLHFVDKTALLDAVCGRYFEKLDEEMQ--EVAGAAATALDV
Mb0672c|M.bovis_AF2122/97 AAGTSTMAVYTHFGGMRGLIAAIAEEGLRQFDVALT--VPQTADP--VAD
Rv0653c|M.tuberculosis_H37Rv AAGTSTMAVYTHFGGMRGLIAAIAEEGLRQFDVALT--VPQTADP--VAD
MSMEG_5307|M.smegmatis_MC2_155 EAGVQPPTIYRQFGDKDGLLDAVTRHVLESYIDRKR--LLTDPDADPALI
TH_4549|M.thermoresistible__bu AAGVQPPTIYRHFGDMRRLIDDAASLGMAEYLKAKRERALTD---DPVHD
MAB_2061c|M.abscessus_ATCC_199 AAGVSGPAVYRHFQNKDAILRELCDAAMTELLAGAR--RATT-AGTPTEV
MAV_1669|M.avium_104 SAGVSGPAIYRHFPNKESLLVELLVGISTRLLAGAR--QVRARSADAAAA
Mflv_0941|M.gilvum_PYR-GCK AAGVAAPTLYHHFGTKEGLLEAVVTEAFSDYLERKR---AVPDSGDLVAD
.*. ::* :* ::
MMAR_4304|M.marinum_M LRAQGLAYVRFALQTPELYRLATMGEW--RSGSNVDS-----------AL
MUL_0990|M.ulcerans_Agy99 LRAQGLAYVRFALQTPELYRLATMGEW--RSGSNVDS-----------AL
Mvan_4572|M.vanbaalenii_PYR-1 LRAQGMAYVRFALKNPVLYRIAMMGPG--SPGSDVDV-----------AL
Mb0672c|M.bovis_AF2122/97 LLAIGTAYRRYAIERPHMYRLMFGSTS--AHGINVPARDVLTLKVAEIEH
Rv0653c|M.tuberculosis_H37Rv LLAIGTAYRRYAIERPHMYRLMFGSTS--AHGINVPARDVLTLKVAEIEH
MSMEG_5307|M.smegmatis_MC2_155 LRELWDLHVEFGMQQPHCYLLTYGQGR---RTSAADETVSILGEVIGRLG
TH_4549|M.thermoresistible__bu LRDGWDLHVEFGLRHPATYTVLYGDPRPDAKPRAVAEGDAILRSLMQRVA
MAB_2061c|M.abscessus_ATCC_199 LHALVDLHAAFGARRRGLLAVYA--------------------------R
MAV_1669|M.avium_104 LDGLIDFHLDFALNEPDLIRIQD--------------------------R
Mflv_0941|M.gilvum_PYR-GCK FAAGWDMHIEFGVQNPVLYRLMFGQAE-ERRSSARDIAETELRRRLQLLA
: : :. . :
MMAR_4304|M.marinum_M DSSAFRHMCASVQAMMDEGIYR-------ADDPTTIALELWTAAHGVAAL
MUL_0990|M.ulcerans_Agy99 DSSAFRHMCASVQAMMDEGIYR-------ADDPTTIALELWTAAHGVAAL
Mvan_4572|M.vanbaalenii_PYR-1 NSSAFGHLLMSVQALIAEGVYP-------QADPTMLALELWTAVHGIAAM
Mb0672c|M.bovis_AF2122/97 QHPSFAHVVRAVHRCLLAGRFATALGADDDTAIVATAAQFWSQIHGFVML
Rv0653c|M.tuberculosis_H37Rv QHPSFAHVVRAVHRCLLAGRFATALGADDDTAIVATAAQFWSQIHGFVML
MSMEG_5307|M.smegmatis_MC2_155 AAGRLTMSVRRATDYFRASGTGFVMSQLSLASEERDAELSGIIFEATLAA
TH_4549|M.thermoresistible__bu EAGRLRIGVDRAAEMMHSACRGVTLTLISQTPAERDPELSAATREAMLAA
MAB_2061c|M.abscessus_ATCC_199 EHRSLSPLATRALRRRQHSYESFWVEALVRARGDLDRERAQGLVAAVLSL
MAV_1669|M.avium_104 DLAYLPKPAERQVRKAQRQYVEVWVGVLRELNPELAEADARLTAHAVFGL
Mflv_0941|M.gilvum_PYR-GCK DEGLLRIDVDDAVSVTTGMAFG-CVTQLIHSGGDATSAVAQAIRHALITE
: .
MMAR_4304|M.marinum_M LIAKPHLPFGDVAAFADRVLGAVLCGHMVAGLVGPQATSRDLMDWVVQHR
MUL_0990|M.ulcerans_Agy99 LIAKPHLPFGDVAAFADRVLGAVLCGHMVAGLVGPQATSRDLMDWVVQHR
Mvan_4572|M.vanbaalenii_PYR-1 LISRPYLPWGGAEEFADRLMRAVCCGQALSVAIRPDESPREIMTWLKEVV
Mb0672c|M.bovis_AF2122/97 ELAG---FYGDRGAAVEPVLAAMTVNLLVALGDSPERAQCSLRAEQTQKN
Rv0653c|M.tuberculosis_H37Rv ELAG---FYGDRGAAVEPVLAAMTVNLLVALGDSPERAQCSLRAEQTQKN
MSMEG_5307|M.smegmatis_MC2_155 VSTDGRRGRG-RKANDVRARAVALREALPESRTPALSAAEQGLLAEWLDR
TH_4549|M.thermoresistible__bu ITTGGAAGPA-DGAG-VATRAIALKSVLETGGAGVLTEAEAELLKEWLDR
MAB_2061c|M.abscessus_ATCC_199 LNASAYMPASLDDATIEAMLAAAAVAALFSRAVTQQVDGAPHRI------
MAV_1669|M.avium_104 LNSTPHSMKSQDSGRGKPARAARSRAIMRAMTVAALAAGNQCP-------
Mflv_0941|M.gilvum_PYR-GCK LTGSPVRHDDPAQAARQVLAGLGTMPALFTPAEEGLLRQWLRTVAEHLDT
MMAR_4304|M.marinum_M PSEESRV------
MUL_0990|M.ulcerans_Agy99 PSEESRV------
Mvan_4572|M.vanbaalenii_PYR-1 DERQRG-------
Mb0672c|M.bovis_AF2122/97 TLGRAT-------
Rv0653c|M.tuberculosis_H37Rv TLGRAT-------
MSMEG_5307|M.smegmatis_MC2_155 LADQA--------
TH_4549|M.thermoresistible__bu LSQ----------
MAB_2061c|M.abscessus_ATCC_199 -------------
MAV_1669|M.avium_104 -------------
Mflv_0941|M.gilvum_PYR-GCK PAAPRRTRKKGNR