For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AGTDAGTDAGTEASSTRERILAATAKVLGSNGTTKLSLSDVASQAGVSRPTLYRWFSSKQDLLDAFVVWE RRGYEQAVAQATGALPASERLDAALRVIAEYQRSYPGLRMVDIEPEHVIRRLSQIMPLLRERLQRLTTGP DSDVAAATAVRVAISHYLVRSDDADDFLAQLRHVARPRRRE*
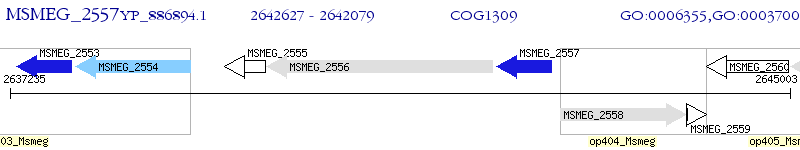
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_2557 | - | - | 100% (182) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_2553 | - | 7e-54 | 59.32% (177) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_0676 | - | 2e-08 | 29.73% (148) | putative transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1998c | - | 7e-08 | 34.02% (97) | transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_4112 | - | 2e-66 | 74.23% (163) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1963c | mce3R | 7e-08 | 34.02% (97) | transcriptional repressor (probably TETR-family) MCE3R |
| M. leprae Br4923 | MLBr_00815 | - | 4e-05 | 27.84% (97) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_2046 | - | 4e-08 | 41.94% (62) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0389 | - | 2e-53 | 63.75% (160) | putative regulatory protein |
| M. avium 104 | MAV_5134 | - | 3e-55 | 63.75% (160) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_0276 | - | 5e-65 | 74.53% (161) | transcriptional regulator, TetR family protein |
| M. ulcerans Agy99 | MUL_1039 | - | 3e-53 | 63.12% (160) | putative regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_2233 | - | 8e-68 | 73.81% (168) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1998c|M.bovis_AF2122/97 MASVAQPVRRRPKDRKKQILDQAVGLFIERGFHSVKLEDIAEAAGVTARA
Rv1963c|M.tuberculosis_H37Rv MASVAQPVRRRPKDRKKQILDQAVGLFIERGFHSVKLEDIAEAAGVTARA
Mflv_4112|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2233|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2557|M.smegmatis_MC2_155 --------------------------------------------------
TH_0276|M.thermoresistible__bu --------------------------------------------------
MMAR_0389|M.marinum_M --------------------------------------------------
MUL_1039|M.ulcerans_Agy99 --------------------------------------------------
MAV_5134|M.avium_104 --------------------------------------------------
MAB_2046|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00815|M.leprae_Br4923 --------------------------------------------------
Mb1998c|M.bovis_AF2122/97 LYRHYDNKQALLAEAIRTGQDQYQSARRLTEGETEPTPRPLNADLEDLIA
Rv1963c|M.tuberculosis_H37Rv LYRHYDNKQALLAEAIRTGQDQYQSARRLTEGETEPTPRPLNADLEDLIA
Mflv_4112|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2233|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2557|M.smegmatis_MC2_155 --------------------------------------------------
TH_0276|M.thermoresistible__bu --------------------------------------------------
MMAR_0389|M.marinum_M --------------------------------------------------
MUL_1039|M.ulcerans_Agy99 --------------------------------------------------
MAV_5134|M.avium_104 --------------------------------------------------
MAB_2046|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00815|M.leprae_Br4923 --------------------------------------------------
Mb1998c|M.bovis_AF2122/97 AAVASRALTVLWQREARYLNEDDRTAVRRRINAIVAGMRDSVLLEVPDLS
Rv1963c|M.tuberculosis_H37Rv AAVASRALTVLWQREARYLNEDDRTAVRRRINAIVAGMRDSVLLEVPDLS
Mflv_4112|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2233|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2557|M.smegmatis_MC2_155 --------------------------------------------------
TH_0276|M.thermoresistible__bu --------------------------------------------------
MMAR_0389|M.marinum_M --------------------------------------------------
MUL_1039|M.ulcerans_Agy99 --------------------------------------------------
MAV_5134|M.avium_104 --------------------------------------------------
MAB_2046|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00815|M.leprae_Br4923 --------------------------------------------------
Mb1998c|M.bovis_AF2122/97 PQHSELRAWAVSSTLTSLGRHSLSLPGEELKKLLYQACMAAARTPPVCEL
Rv1963c|M.tuberculosis_H37Rv PQHSELRAWAVSSTLTSLGRHSLSLPGEELKKLLYQACMAAARTPPVCEL
Mflv_4112|M.gilvum_PYR-GCK ------------------------------------------------MT
Mvan_2233|M.vanbaalenii_PYR-1 ------------------------------------------------MP
MSMEG_2557|M.smegmatis_MC2_155 ------------------------------------------------MA
TH_0276|M.thermoresistible__bu ------------------------------------------------MP
MMAR_0389|M.marinum_M ------------------------------------------------MS
MUL_1039|M.ulcerans_Agy99 ------------------------------------------------MS
MAV_5134|M.avium_104 ------------------------------------------------MS
MAB_2046|M.abscessus_ATCC_1997 ------------------------------------------------MP
MLBr_00815|M.leprae_Br4923 -----------------------------MNNLAKAAQRRTVRPIDRGRL
Mb1998c|M.bovis_AF2122/97 PPLPAGDAARDEADVLFSRYETLLAAGARLFRAQGYPAVNTSEIGKGAGI
Rv1963c|M.tuberculosis_H37Rv PPLPAGDAARDEADVLFSRYETLLAAGARLFRAQGYPAVNTSEIGKGAGI
Mflv_4112|M.gilvum_PYR-GCK MPSP---APGSAD-PDNPTRQRILAATAEVLGRNGTTRLSLSEVAAQAGV
Mvan_2233|M.vanbaalenii_PYR-1 QPPPGDRGPQSADDPDSPTRQRILAATAEVLGRNGTTKLSLSEVASQAGV
MSMEG_2557|M.smegmatis_MC2_155 GTDAG----TDAGTEASSTRERILAATAKVLGSNGTTKLSLSDVASQAGV
TH_0276|M.thermoresistible__bu ------------DRIAASARERILTATAEVLGRAGMTKLSLSDVAQQAGV
MMAR_0389|M.marinum_M QAFG----AIDTTDGDTSTRQRILAATAEVLARSGKTKLSLSEVAAQAGV
MUL_1039|M.ulcerans_Agy99 QAFG----AIDTTDGDTSMRQRILAATAEVLARSGKTKLSLSEVAAQAGV
MAV_5134|M.avium_104 HPVR----ATAIADTDTSTRQRILAATAEVLGRNGKTKLSLSDVATQAGV
MAB_2046|M.abscessus_ATCC_1997 TEPVR-------VSFRRHVREKVLRATRELAIEKGWDQVRMSEVAESVGV
MLBr_00815|M.leprae_Br4923 SDDIATPNQRSNRLSRDKRRGQLLIVASDVFVDHGYHAAGMDEIADRAGV
:* . : * .::. .*:
Mb1998c|M.bovis_AF2122/97 AGPGLYRSFSSKQAILDALIRRLDEWRCLECIRALRANQQAAQR--LRGL
Rv1963c|M.tuberculosis_H37Rv AGPGLYRSFSSKQAILDALIRRLDEWRCLECIRALRANQQAAQR--LRGL
Mflv_4112|M.gilvum_PYR-GCK SRPTLYRYFADKRELLDVFVVWERQY-YERAIAKATAELPAHER--LDAA
Mvan_2233|M.vanbaalenii_PYR-1 SRPTLYRYFADKRELLDVFVVWERQY-YERAIARATADLPVSER--LDAA
MSMEG_2557|M.smegmatis_MC2_155 SRPTLYRWFSSKQDLLDAFVVWERRG-YEQAVAQATGALPASER--LDAA
TH_0276|M.thermoresistible__bu SRPTLYRWFSGKEELLDAFAQWERQM-YERALAEATAGLPAEER--LDAV
MMAR_0389|M.marinum_M SRPTLYRWFASKEELLATFSRYERQV-FESGLSKATAGLKGVDK--LDAA
MUL_1039|M.ulcerans_Agy99 SRPTLYRWFASKEELLATFSRYERQV-FESGLSKATAGLKGVDK--LDAA
MAV_5134|M.avium_104 SRPTLYRWFASKEELLSAFSAYERQI-FESGLVKATAGLKGVDK--LDAV
MAB_2046|M.abscessus_ATCC_1997 SRPTLYKEFGDKQGLGDALVVSEGQR-FMEGILAVLAEHVGDVRGGITAA
MLBr_00815|M.leprae_Br4923 SKPVLYQHFSSKLELYLAVLERHMNNLVSSVQQALRTTTNNRQR--LHAA
: * **: *..* : .. . : : .
Mb1998c|M.bovis_AF2122/97 VQGHVRISLDAPDLVAVSVTELSHASVEVRDGYLRNQGDREAVWIDLIG-
Rv1963c|M.tuberculosis_H37Rv VQGHVRISLDAPDLVAVSVTELSHASVEVRDGYLRNQGDREAVWIDLIG-
Mflv_4112|M.gilvum_PYR-GCK LRVIVEYQLSYPGMRMVDVEPD--------------------QVIRRMS-
Mvan_2233|M.vanbaalenii_PYR-1 LRVIVEYQLSYPGLRMVDVEPD--------------------QVIKRLT-
MSMEG_2557|M.smegmatis_MC2_155 LRVIAEYQRSYPGLRMVDIEPE--------------------HVIRRLS-
TH_0276|M.thermoresistible__bu LRVIVEYQQSYPGLRMVDIEPA--------------------HLIGRLS-
MMAR_0389|M.marinum_M LRFIVDYQYSYSGVRMVDIEPE--------------------LVISQMS-
MUL_1039|M.ulcerans_Agy99 LRFIVDYQYSYSGVRMVDIEPE--------------------LVISQMS-
MAV_5134|M.avium_104 LRFIVDYQHSYSGVRMVDVEPE--------------------HTIAQFS-
MAB_2046|M.abscessus_ATCC_1997 VQFTLCEAEDSPLLKAVLTSNPSGNDRGGSSSTGVLPLLPTSASLLQLCS
MLBr_00815|M.leprae_Br4923 VQAFFDFIEHDGQGYRLIFENDNITEPRVAAQVRVATESCIDAVFALIS-
:: : : :
Mb1998c|M.bovis_AF2122/97 -KLVPATSVAQGRLLVAAAISFIEDVARTWHLTRYAGVADEISGLALAIL
Rv1963c|M.tuberculosis_H37Rv -KLVPATSVAQGRLLVAAAISFIEDVARTWHLTRYAGVADEISGLALAIL
Mflv_4112|M.gilvum_PYR-GCK -RVLPLMRERLERLCTGPDAALSAATAVRVAISQYLVRSDDSA-DFLAQL
Mvan_2233|M.vanbaalenii_PYR-1 -RVLPLMRDRLQRLCTGPDAALAAATAVRVAISQYLVRSDDSE-QFLAQL
MSMEG_2557|M.smegmatis_MC2_155 -QIMPLLRERLQRLTTGPDSDVAAATAVRVAISHYLVRSDDAD-DFLAQL
TH_0276|M.thermoresistible__bu -AILPVMRDRLERLCTGPTASAAAATAVRVAVSHYLVQSDDAG-DFLAQL
MMAR_0389|M.marinum_M -GVIPEMREGLQRLLPGPNAAVKAATAIRIAISHYIVRSDDAD-QFLAQL
MUL_1039|M.ulcerans_Agy99 -SVIPEMREGLQRLLPGPNAAVKAATAIRIAISHYIVRSDDAD-QFLAQL
MAV_5134|M.avium_104 -WVIPQMREGLQRHLPGPNAAVKAATVIRIAISHYIVRSDDAD-QFLAQL
MAB_2046|M.abscessus_ATCC_1997 AALITWFNDHFDDLDP-EDVEEVADVLVRLTVSHVVLPAADIATTGERIS
MLBr_00815|M.leprae_Br4923 --ADSRLDPHRARMVAVGLVGISVDCARYWLDTNRPISKSDAVEGTVQFA
. :. :
Mb1998c|M.bovis_AF2122/97 TSGAGN--------------LLRA-
Rv1963c|M.tuberculosis_H37Rv TSGAGN--------------LLRA-
Mflv_4112|M.gilvum_PYR-GCK RHAARV--------------RHPGR
Mvan_2233|M.vanbaalenii_PYR-1 RHAARV--------------KHRDM
MSMEG_2557|M.smegmatis_MC2_155 RHVARP--------------RRRE-
TH_0276|M.thermoresistible__bu RHAVGLPIAPGAVSAKTPLQPRRNP
MMAR_0389|M.marinum_M RHAVGL--------------KGG--
MUL_1039|M.ulcerans_Agy99 RHAVGL--------------KGG--
MAV_5134|M.avium_104 RHAVGI--------------KAPTD
MAB_2046|M.abscessus_ATCC_1997 RVALRYLG------------VVHSL
MLBr_00815|M.leprae_Br4923 WGGLSHVP------------LTRL-