For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TQRVAIVTGASGGIGHDVAVRLARAGMAVAVHYAGNRDRAQQTADEITALGGAATVVTADVADEQQVAAL FDEVESEFGGVDVVVNTAGIMTLAPLAELDLADLDRMLRTNVRGTFAVSQQAARRVRAGGAIINFSTSVT KIAAPTYTAYAATKGAVDAMTLILAKEMRGRDVTVNAVAPGPTATPLYFEGKPQEVIDRAKAAPPLERLG EPADIAEIVAFLAGPARWVNGQVIYANGGVV*
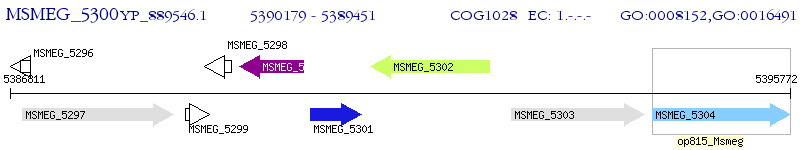
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5300 | - | - | 100% (242) | short-chain type dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_2029 | - | 4e-38 | 40.33% (243) | 3-ketoacyl-ACP/CoA redutase |
| M. smegmatis MC2 155 | MSMEG_4351 | - | 4e-36 | 37.45% (243) | oxidoreductase YjgI |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1963c | - | 8e-32 | 36.93% (241) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1601 | - | 2e-33 | 36.18% (246) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv1928c | - | 8e-32 | 36.93% (241) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 5e-21 | 32.92% (243) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_3426c | - | 8e-36 | 33.06% (242) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_4927 | - | 2e-31 | 35.51% (245) | dehydrogenase/reductase |
| M. avium 104 | MAV_4177 | - | 6e-34 | 38.10% (252) | putative short-chain type dehydrogenase/reductase y4lA |
| M. thermoresistible (build 8) | TH_3122 | - | 3e-35 | 34.47% (235) | PUTATIVE short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_0478 | - | 2e-29 | 34.69% (245) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5843 | - | 1e-35 | 35.15% (239) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1963c|M.bovis_AF2122/97 ------------MSVLDLFDLHGKRALITGASTGIGKRVALAYVEAGAQV
Rv1928c|M.tuberculosis_H37Rv ------------MSVLDLFDLHGKRALITGASTGIGKRVALAYVEAGAQV
Mvan_5843|M.vanbaalenii_PYR-1 ---------------MSTAPLAGKRALVTGGSRGIGAEIVRRLAADGAAV
MMAR_4927|M.marinum_M -------------MG----QFENKVAIVTGAAQGIGQAYADALAREGARV
MUL_0478|M.ulcerans_Agy99 -------------MG----QFENKVAIVTGAAQGIGQAYADALAREGARV
Mflv_1601|M.gilvum_PYR-GCK -------------MGLYGDQFNEKVAIVTGAGGGIGQAYAEALAREGAAV
MAV_4177|M.avium_104 -------------MGLR--GLADKVAVVVGGATGIGAATAARLAGEGCRV
MSMEG_5300|M.smegmatis_MC2_155 --------------------MTQRVAIVTGASGGIGHDVAVRLARAGMAV
MAB_3426c|M.abscessus_ATCC_199 MVDIPHTIDPVQPYRPQEQPLRGKVAVITGASANMGAALARTLAQDGAQV
TH_3122|M.thermoresistible__bu ---------------------------VAAGAKNLGGLISTTLAERGVNV
MLBr_01807|M.leprae_Br4923 --MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRV
:... .:* . * *
Mb1963c|M.bovis_AF2122/97 AIA-ARH--LDALEKLADEIGTSGGKVVPVCCDVSQHQQVTSMLDQVTAE
Rv1928c|M.tuberculosis_H37Rv AIA-ARH--LDALEKLADEIGTSGGKVVPVCCDVSQHQQVTSMLDQVTAE
Mvan_5843|M.vanbaalenii_PYR-1 AFTYAAS--AAHAEKVVAEVAASGGEAVAIRADSADPTQVADAVEQTVAE
MMAR_4927|M.marinum_M VVADIN---AEAAEKTAKQIVADGGTAIHVPVDVSDEESARAMADRTVSE
MUL_0478|M.ulcerans_Agy99 VVADIN---AEAAEKTAKQIVADGGTAIHVPVDVSDEESARAMADRTVSE
Mflv_1601|M.gilvum_PYR-GCK VVADIN---VEGAQKVADGIKGEGGNALAVRVDVSDPDSAKEMAAQALSE
MAV_4177|M.avium_104 VIGDVA---VDAARQTADRIGAAGGTATQVAFDLADPASVATLIDCAATT
MSMEG_5300|M.smegmatis_MC2_155 AVHYAGN--RDRAQQTADEITALGGAATVVTADVADEQQVAALFDEVESE
MAB_3426c|M.abscessus_ATCC_199 VVHYRSDSKAQQAERVVRQIQESGSDAVSFQADLSVPENCTKLADAAFEA
TH_3122|M.thermoresistible__bu AVHYHSAATEPDADKTVAAVEATGARAIKVQGDLTVPANVTRLFDATVEA
MLBr_01807|M.leprae_Br4923 AVTHRAS--------------VVPDWFFGVECDVTDNDAIGAAFKAVEEH
.. . * : .
Mb1963c|M.bovis_AF2122/97 LGGIDIAVCNAGIITVT----PMLDMPLEEFQRLQNTNVTGVFLTAQAAA
Rv1928c|M.tuberculosis_H37Rv LGGIDIAVCNAGIITVT----PMLDMPLEEFQRLQNTNVTGVFLTAQAAA
Mvan_5843|M.vanbaalenii_PYR-1 LGGLDILVNNSAVAHIA----PIDDFPAEEFDRLVAINIGGVYWAVRSAI
MMAR_4927|M.marinum_M FGGIDYLVNNAAIYGGMKLDL-LLTVPLDYYKKFMSVNHDGVLVCTRAVY
MUL_0478|M.ulcerans_Agy99 FGGIDYLVNNAAIYGGMKLDL-QLTVPLDYYKKFMSVNHDGVLVCTRAVY
Mflv_1601|M.gilvum_PYR-GCK FGGIDYLVNNAAIFGGMKLDF-LITVDWDYYKKFMSVNLDGALVCTRAVY
MAV_4177|M.avium_104 YGGVDLLFNVGADMSTIRADTDVVDIDFDVWDRVMTVSLRGYVAAMKYAI
MSMEG_5300|M.smegmatis_MC2_155 FGGVDVVVNTAGIMT---LAP-LAELDLADLDRMLRTNVRGTFAVSQQAA
MAB_3426c|M.abscessus_ATCC_199 FGRWDILINTAGMIVR----KPLADITEDEYDGVFNANAKTVFFMMREAA
TH_3122|M.thermoresistible__bu FGGVDIAVNTVGRVLR----KPIVETTEAEYDAMFDINAKAAYFFLQEAG
MLBr_01807|M.leprae_Br4923 QGPVEVLVANAGITSDM----LIMRMSEEQFQKVIDVNLTGVFRVAKLAS
* : . . . . . : .
Mb1963c|M.bovis_AF2122/97 KAMVKQGQGGVIINTASMSGHIINVPQQVSHYCASKAAVIHLTKAMAVEL
Rv1928c|M.tuberculosis_H37Rv KAMVKQGQGGVIINTASMSGHIINVPQQVSHYCASKAAVIHLTKAMAVEL
Mvan_5843|M.vanbaalenii_PYR-1 RYLS---EGARIINIGSINADRIPGPG-LSVYGMTKGAVSSFTRGLARDL
MMAR_4927|M.marinum_M KHMAKRGGGAIVN---QSSTAAWLYS---NFYGLAKVGVNGLTQQLSREL
MUL_0478|M.ulcerans_Agy99 KHMAKRGGGAIVN---QSSTAAWLYS---NFYGLAKVGVNGLTQQLSREL
Mflv_1601|M.gilvum_PYR-GCK RKMAKRGGGAIVN---QSSTAAWLYS---NFYGLAKVGINGLTQQLATEL
MAV_4177|M.avium_104 PRMLDRGGGAIVN---MSSAAAFQGEPARPAYATAKAGIGALTRHVASRW
MSMEG_5300|M.smegmatis_MC2_155 RRVR--AGGAIIN---FSTSVTKIAAPTYTAYAATKGAVDAMTLILAKEM
MAB_3426c|M.abscessus_ATCC_199 RRMA--DNGRILS---FVTTMVGALAPTYSAYAGSKAPVEHFTKALAKEV
TH_3122|M.thermoresistible__bu KRLN--DGGSVVT---ILTALLAAFTDGYSTYAGAKSPVEHFTRAAAKEF
MLBr_01807|M.leprae_Br4923 RNMIKQRFGRYIF---MGSVVGLSGVPGQANYAASKSGLIGMARSLAREL
: * : . * :* : :: :
Mb1963c|M.bovis_AF2122/97 APHKIRVNSVSPGYILTELV---EPYTEYQPLWEPKIPLGRLGRPEELAG
Rv1928c|M.tuberculosis_H37Rv APHKIRVNSVSPGYILTELV---EPYTEYQPLWEPKIPLGRLGRPEELAG
Mvan_5843|M.vanbaalenii_PYR-1 GPRGITVNNVQPGPIDTDAN---PDDGAFAESLKQVVAQGRYGHTSDVAS
MMAR_4927|M.marinum_M GGMKIRINAIAPGPIDTEATRTVTPG-EFVKDMVKQIPLSRMGTPEDLVG
MUL_0478|M.ulcerans_Agy99 GGMKIRINAIAPGPIDTEATRTVTPG-EFVKDMVKQIPLSRMGTPEDLVG
Mflv_1601|M.gilvum_PYR-GCK GGQNIRVNAIAPGPIDTEANRTTTPQ-EMVADIVKGIPLSRMGQPDDLVG
MAV_4177|M.avium_104 GKDNIRCNAVAPGFTATETIRSVPQWPELEAAALKRIRGPRVGDPADVAA
MSMEG_5300|M.smegmatis_MC2_155 RGRDVTVNAVAPGPTATPLYFEGKPQ-EVIDRAKAAPPLERLGEPADIAE
MAB_3426c|M.abscessus_ATCC_199 GGRGITANCIAPGPLQTSFFYPAESD-ENIAWLESMSINGQIGRGDDILP
TH_3122|M.thermoresistible__bu APRQISVNNVAPGPMDTPFFYGQETP-ERVTFHKSQGLGNRLTRIEDIAP
MLBr_01807|M.leprae_Br4923 GSRSITANVVAPGFIDTEMTRAMDSK--YQEAALQAIPLGRIGAVEEIAG
: * : ** * : ::
Mb1963c|M.bovis_AF2122/97 LYLYLASEASSYMTGSDIVIDGGYTCP--
Rv1928c|M.tuberculosis_H37Rv LYLYLASEASSYMTGSDIVIDGGYTCP--
Mvan_5843|M.vanbaalenii_PYR-1 LVSFLAGPESGYITGAHLNVDGGFTV---
MMAR_4927|M.marinum_M MCLFLLGDQASWITGQIFNVDGGQIIRS-
MUL_0478|M.ulcerans_Agy99 MCLFLLGDQASRITGQIFNVDGGQIIRS-
Mflv_1601|M.gilvum_PYR-GCK MLLFLLSDQAKWVTGQIFNVDGGQIIR--
MAV_4177|M.avium_104 LVAFLLSAEGDWINGQVINIDGGTVLR--
MSMEG_5300|M.smegmatis_MC2_155 IVAFLAG-PARWVNGQVIYANGGVV----
MAB_3426c|M.abscessus_ATCC_199 VARLLVLPESGWTTAQTIFVNGGIISTIN
TH_3122|M.thermoresistible__bu LVAFLTD-EGHWITGQTIFANGGYTTR--
MLBr_01807|M.leprae_Br4923 AVSFLASEDAGYISGAVIPVDGGLGMGH-
* . .. : :**