For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTTDATARTRVSLTGVSETALLTLQVRANEARRPDGLIKDPMAVQLVDSIDFDFAKFGHSHRQDMALRSL IFDRATSDYLRDHPKATVVALAEGLQTSFYRLNEAGVGDQFCWLTVDLPPMVEIRNKLLPPSDRVRTCAQ SALDYSWMDQVDDSEGVFITAEGLLMYLQPEESMGLIKECARRFPGGQMIFDLPPSWFAAWARKGMRTSR RYRVPPMPFSLSPAEVADLVNTVPGVRVVHDLPFPPARGKFMRTVVWGVQRLPLFDPIRPVMALLEFG
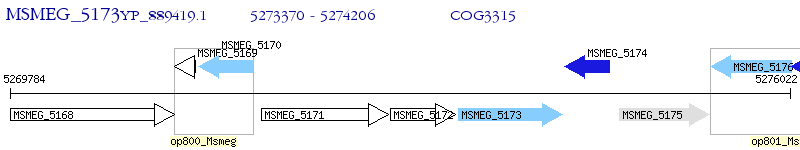
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5173 | - | - | 100% (278) | O-methyltransferase, putative |
| M. smegmatis MC2 155 | MSMEG_6661 | - | 8e-86 | 59.55% (267) | O-methyltransferase, putative |
| M. smegmatis MC2 155 | MSMEG_6473 | - | 1e-25 | 29.66% (236) | tetracenomycin polyketide synthesis O-methyltransferase TcmP |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1184c | omt | 3e-99 | 65.34% (277) | O-methyltransferase |
| M. gilvum PYR-GCK | Mflv_2136 | - | 1e-110 | 72.00% (275) | O-methyltransferase domain-containing protein |
| M. tuberculosis H37Rv | Rv1153c | omt | 1e-100 | 65.70% (277) | O-methyltransferase |
| M. leprae Br4923 | MLBr_02640 | - | 6e-07 | 27.94% (204) | hypothetical protein MLBr_02640 |
| M. abscessus ATCC 19977 | MAB_1292c | - | 8e-75 | 53.56% (267) | O-methyltransferase OMT |
| M. marinum M | MMAR_4300 | omt_2 | 4e-99 | 64.36% (275) | O-methyltransferase Omt_2 |
| M. avium 104 | MAV_2991 | - | 1e-23 | 32.31% (195) | tetracenomycin polyketide synthesis O-methyltransferase TcmP |
| M. thermoresistible (build 8) | TH_0460 | omt | 1e-112 | 73.31% (266) | PROBABLE O-METHYLTRANSFERASE OMT |
| M. ulcerans Agy99 | MUL_3020 | omt | 2e-82 | 58.87% (265) | O-methyltransferase Omt |
| M. vanbaalenii PYR-1 | Mvan_4568 | - | 1e-107 | 66.22% (299) | O-methyltransferase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2136|M.gilvum_PYR-GCK --MSPTDKHSA-DLTGVSETALMTLLVRATEARRPDSLIDDPMAVKLVDS
Mvan_4568|M.vanbaalenii_PYR-1 --MNHTGKHTATDVTGVSETALLTLLVRATEARRPDSLIDDPMAIHLVDS
TH_0460|M.thermoresistible__bu ---VSSGPLAKPELTGVSETALLTLLVRATEARRPDGIIDDPMAVRLVDS
MSMEG_5173|M.smegmatis_MC2_155 -MTTDATARTRVSLTGVSETALLTLQVRANEARRPDGLIKDPMAVQLVDS
Mb1184c|M.bovis_AF2122/97 MS-AHKPAKQRVALTGVSETALLTLNARAAEARRRDTIIDDPMAVALVES
Rv1153c|M.tuberculosis_H37Rv MS-AHKPAKQRVALTGVSETALLTLNARAAEARRRDAIIDDPMAVALVES
MMAR_4300|M.marinum_M MTPADEFAQQKVALTGVSETALLTLNARAAEARRRDPIIEDPMAVRLVDS
MUL_3020|M.ulcerans_Agy99 -MGHSDNNVNAGMLGGVSETALLTLNGRAHQAAKPNPIIDDPMAIKLVES
MAB_1292c|M.abscessus_ATCC_199 --MAYMAAIDARHLDGVSETALITLNQRATEANRPDGVLEDPMAIVLRDS
MAV_2991|M.avium_104 MTATEKVDFSSVAWGSVEWTNLVTLYLRACESRSRQPILGDTAAAEAVDR
MLBr_02640|M.leprae_Br4923 ---MRTHDDTWDIKTSVGTTAVMVAAARAAETDRPDALIRDPYAKLLVTN
.* * ::. ** :: : :: *. *
Mflv_2136|M.gilvum_PYR-GCK L--------DFDRAKFG-----FVRRQDMAVRALVFDKWTRRYLVDHPRA
Mvan_4568|M.vanbaalenii_PYR-1 I--------DFDFAKFG-----YVRRQDMSLRALAFDKQTRRYLRDHPKA
TH_0460|M.thermoresistible__bu I--------DFDFAKFG-----PSRRQDMALRAKAFDTATRRYLRAHPKA
MSMEG_5173|M.smegmatis_MC2_155 I--------DFDFAKFG-----HSHRQDMALRSLIFDRATSDYLRDHPKA
Mb1184c|M.bovis_AF2122/97 I--------DFGFAKFG-----PTG-QGFALRARAFDMAAQHYLDQHPAA
Rv1153c|M.tuberculosis_H37Rv I--------DFDFAKFG-----PTG-QGFALRARAFDMAAQHYLDQHPAA
MMAR_4300|M.marinum_M I--------DFDFAKFG-----PTR-QDIALRALAFDTRALAYLNKHPSA
MUL_3020|M.ulcerans_Agy99 N--------DFDFEKFG-----RRG-QEMALRSLAVDRCEIAYLTKHPGA
MAB_1292c|M.abscessus_ATCC_199 L--------DYDYHHFG-----RTH-QGIALRALTFDTVSNEYLAEHPRA
MAV_2991|M.avium_104 I--------RYDFKRIHRMSLPASNQYLVALRAVQFDDWCAEFLARHPDA
MLBr_02640|M.leprae_Br4923 TGAGALWEAMLDPSMVAKVEAIDAEAAAMVEHMRSYQAVRTNFFDTYFNN
. . . : : :: :
Mflv_2136|M.gilvum_PYR-GCK TVVA-------LAEGLQTSFYRLDSGPESVGHDFRWLTVDLPPMIDLRTK
Mvan_4568|M.vanbaalenii_PYR-1 TVVA-------LAEGLQTSFYRLDAQGPDVGHDFRWLTVDLPPMIELRRK
TH_0460|M.thermoresistible__bu TVVA-------LAEGLQTSFFRLN--AADVGDEFRWVTVDLAPVIELRRQ
MSMEG_5173|M.smegmatis_MC2_155 TVVA-------LAEGLQTSFYRLN--EAGVGDQFCWLTVDLPPMVEIRNK
Mb1184c|M.bovis_AF2122/97 TVVA-------LAEGLQTSFWRLD--VAIPGGQFRWLTVDLPPIVDLRTR
Rv1153c|M.tuberculosis_H37Rv TVVA-------LAEGLQTSFWRLD--VAIPGGQFRWLTVDLPPIVDLRTR
MMAR_4300|M.marinum_M TVVA-------LAEGLQTSFWRLD--SALPGKKFRWLTVDLPPIIDIRKR
MUL_3020|M.ulcerans_Agy99 TVVA-------HAEGFQTGFWRVS--NALPDTQFRWVSLDLEPVMELRRR
MAB_1292c|M.abscessus_ATCC_199 TVVA-------LAEGLQTSFWRID------NGELNWLSVDLEPIVRLRRQ
MAV_2991|M.avium_104 VVLH-------LGCGLDGRAFRVS-----VPPSLSWFDVDQPSVIGLRRR
MLBr_02640|M.leprae_Br4923 AVIDGIRQFVILASGLDSRAYRLD-----WPTGTTVYEIDQPKVLAYKST
.*: . *:: :*:. :* :: :
Mflv_2136|M.gilvum_PYR-GCK LLPPSDRVRTLAQS----ALDFSWMDQVDTSEG-----------------
Mvan_4568|M.vanbaalenii_PYR-1 LLPASDRVQMLAQS----ALDFSWMDRVPTDDGDVNQAGDDGDVNQAGDD
TH_0460|M.thermoresistible__bu LLPADDRVTMLAQS----ALDYSWMDRIDTSEG-----------------
MSMEG_5173|M.smegmatis_MC2_155 LLPPSDRVRTCAQS----ALDYSWMDQVDDSEG-----------------
Mb1184c|M.bovis_AF2122/97 LLPSSPRVSVCAQS----ALDYSWMDSVDPAGG-----------------
Rv1153c|M.tuberculosis_H37Rv LLPSSPRVSVCAQS----ALDYSWMDSVDPAGG-----------------
MMAR_4300|M.marinum_M LLPNSPRVSVCAQS----ALDYGWMDSVDTSGG-----------------
MUL_3020|M.ulcerans_Agy99 LLPDSARVTNLAQS----ALDYTWMDKVDSCAA-----------------
MAB_1292c|M.abscessus_ATCC_199 LLPPSDRVRYVAQS----ALDHSWMDQVDDSNG-----------------
MAV_2991|M.avium_104 LYDDTENYRMIGSS----VTELQWLDEVPAGRP-----------------
MLBr_02640|M.leprae_Br4923 TLAEHGVTPTADRREVPIDLRQDWPPALRSAGFDPSAR------------
* :
Mflv_2136|M.gilvum_PYR-GCK ----------VFITAEGLLMYLQPAEAMSLITECAARFPGGQMIFDLPPS
Mvan_4568|M.vanbaalenii_PYR-1 GDVDQAGADGVFITAEGLLMYLQPEESMALITECAARFPGGRMMFDLPPS
TH_0460|M.thermoresistible__bu ----------VFITAEGLLMYLQPDEAMGLIRECARRFPGGQMMFDLPPS
MSMEG_5173|M.smegmatis_MC2_155 ----------VFITAEGLLMYLQPEESMGLIKECARRFPGGQMIFDLPPS
Mb1184c|M.bovis_AF2122/97 ----------VFITAEGLLMYLQPEQALGLIAQCAQTFPGGQMLFDLPPR
Rv1153c|M.tuberculosis_H37Rv ----------VFITAEGLLMYLQPEQALGLIAQCAQTFPGGQMLFDLPPR
MMAR_4300|M.marinum_M ----------VFITAEGLLMYLEPDQALDLIAQCAKRFPGAQMLFDLPPR
MUL_3020|M.ulcerans_Agy99 ----------VFITAEGLLMYPQPNQAMELISQCAKRFAGGQMFFDLPPT
MAB_1292c|M.abscessus_ATCC_199 ----------VLITAEGLFMYLERDVVFDLIAACAKRFPGGWMVFDTIPW
MAV_2991|M.avium_104 ----------TLMVAEGLLMYLREDEVRRLLQRLIDRFPGGEMHFDTLSA
MLBr_02640|M.leprae_Br4923 ----------TAWLAEGLLMYLPATAQDGLFTEIGGLSAVGSRIAVETSP
. ****:** *: . . .
Mflv_2136|M.gilvum_PYR-GCK AFAFLVG-RGVRASLRYRVPPMPFSLTIAEAADLVNTVPGVRAVHDLPAE
Mvan_4568|M.vanbaalenii_PYR-1 AFAPLVR-RGFRASLRYRVPPMPFTLSPAEAADLVNTVPGVRAVHDVPAE
TH_0460|M.thermoresistible__bu FFAWLTR-RGFRTSLRYKVPPMPFTLSPEQAADLVNTVPGVQAVHDVPIP
MSMEG_5173|M.smegmatis_MC2_155 WFAAWAR-KGMRTSRRYRVPPMPFSLSPAEVADLVNTVPGVRVVHDLPFP
Mb1184c|M.bovis_AF2122/97 WFAGWSR-LGLRTSLRYKVPRMPFSMSVAQAADLVNKVPGVVAVRDLRVP
Rv1153c|M.tuberculosis_H37Rv WFAGWSR-LGLRTSLRYKVPRMPFSMSVAQAADLVNKVPGVVAVRDLRVP
MMAR_4300|M.marinum_M WFSLFTR-LGMRTSIRYKIPPMPFSLSVAQAADLVNTIPGIRAVRDVQLP
MUL_3020|M.ulcerans_Agy99 IVKRVAP-KGMRASRRYRVPPMPFSLSVNQLARLVDTVPGIRAVHDVPMP
MAB_1292c|M.abscessus_ATCC_199 LMSVYSRRRGWRLSEHYTVPPMPFSFTAHQYGQLR-ALDGVSAVREVRMP
MAV_2991|M.avium_104 LAPLLSK----------VFTRGIITWGIRDARRIADWNPRLRFLEQSPAL
MLBr_02640|M.leprae_Br4923 LHGDEWR-EQMQLRFRRVSDALGFEQAVD-VQELIYHDENRAVVADWLNR
: : : :
Mflv_2136|M.gilvum_PYR-GCK HARGRLINALLWTAQRVPLLDP----IRPMFTLLEFG-------
Mvan_4568|M.vanbaalenii_PYR-1 HARGRILNALLWTAQRVPLLDP----VRPMFTLLEFG-------
TH_0460|M.thermoresistible__bu RGRGVLVNTLIWAAQRTPLFDN----VRGTYTLLEFGPPNSAST
MSMEG_5173|M.smegmatis_MC2_155 PARGKFMRTVVWGVQRLPLFDP----IRPVMALLEFG-------
Mb1184c|M.bovis_AF2122/97 PGRGLWVNMALSTVYRLPVFDP----LRPCLTLLEFSRPARG--
Rv1153c|M.tuberculosis_H37Rv PGRGLWVNMALSTVYRLPVFDP----LRPCLTLLEFSRPARG--
MMAR_4300|M.marinum_M AGRGLMFNAALSTVYRLPALDS----LRPCLTLLEFG-------
MUL_3020|M.ulcerans_Agy99 EGRGLFFRTLYPALWRFPPIKP----FRGAYVLLEFD-------
MAB_1292c|M.abscessus_ATCC_199 AGRGTFLKLATPLFYNLPLSDR----IRPAMTVVEFG-------
MAV_2991|M.avium_104 AGYQRIPSTVVRSIYRLTWLTG----GRGYDVLNRFEY------
MLBr_02640|M.leprae_Br4923 HGWRATAQSAPDEMRRVGRWGDGVPMADDKDAFAEFVTAHRL--
. . .. .*