For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSGRVEGKVAFVTGAARGQGRSHALRLAQEGADIIAIDICGPIRPGAETAIPASTSDDLAETANLVKGLD RRVVTAEVDVRDAAAIKAAVDSGVEQLGRLDIIVANAGIGNGGDTLDQTSEEDWDEMIDINLSGVWKSVK AGVPHIVAGGRGGSIVLTSSVGGLKAYPHCGNYVAAKHGVVGLMRGFAVELGQHNIRVNTVHPTHVATPM LHNEGTFKMFRPDLENPGPDDMAPICQMFHTLPIPWVEPIDISNAVLFLASDEARYITGVTLPVDAGSCL K
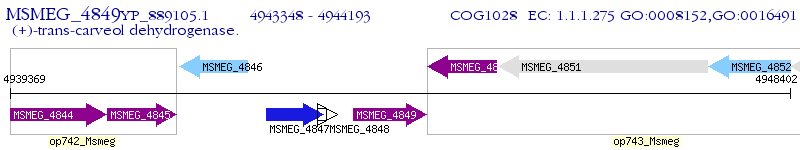
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4849 | - | - | 100% (281) | carveol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6031 | - | e-123 | 75.44% (281) | carveol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2204 | - | 4e-66 | 47.12% (278) | carveol dehydrogenase ((+)-trans-carveol dehydrogenase) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2771 | - | 7e-67 | 49.10% (277) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_1589 | - | 1e-124 | 75.80% (281) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2750 | - | 7e-67 | 49.10% (277) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 9e-22 | 33.66% (202) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0072c | - | 1e-79 | 53.96% (278) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_4754 | - | 1e-136 | 83.27% (281) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_0896 | - | 1e-145 | 88.30% (282) | carveol dehydrogenase ((+)-trans-carveol dehydrogenase) |
| M. thermoresistible (build 8) | TH_3237 | - | 1e-127 | 77.94% (281) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3392 | fabG | 9e-62 | 45.20% (281) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_5168 | - | 1e-126 | 77.22% (281) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2771|M.bovis_AF2122/97 ---MIDRPLEGKVAFITGAARGLGRAHAVR---LAADGANIIAVDICEQI
Rv2750|M.tuberculosis_H37Rv ---MIDRPLEGKVAFITGAARGLGRAHAVR---LAADGANIIAVDICEQI
MUL_3392|M.ulcerans_Agy99 MSRPLDGPLSGRVAFITGAARGQGRAHALR---LARDGADVIAVDLCDQI
MSMEG_4849|M.smegmatis_MC2_155 ----MSGRVEGKVAFVTGAARGQGRSHALR---LAQEGADIIAIDICGPI
MAV_0896|M.avium_104 ----MTGRVEGKVAFVTGAARGQGRSHAVR---LAQEGADIIAVDICKPI
MMAR_4754|M.marinum_M MTEQLGGRVAGKVAFITGAARGQGRSHAVR---LAQEGADIIAVDVCKPI
TH_3237|M.thermoresistible__bu ----MAGRLEGKVAFITGAARGQGRAHAVR---MAQEGADIIAVDICRQI
Mflv_1589|M.gilvum_PYR-GCK ----MGGRVEGKVAFITGAARGQGRSHAVR---LAEEGADIIAIDVCGPI
Mvan_5168|M.vanbaalenii_PYR-1 ----MGGRVEGKVAFITGAARGQGRSHAVR---LAEEGADIIAIDVCGPI
MAB_0072c|M.abscessus_ATCC_199 ----MTGRLTGKVALVTGAARGIGRAQAVR---FAQEGADIIALDICAPV
MLBr_01807|M.leprae_Br4923 ---------MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRL
.* ..** * *:. : :. * *.: :. :
Mb2771|M.bovis_AF2122/97 ASV----PYPLSTADDLAATVELVEDAGGGIVARQGDVRDRASLSVALQA
Rv2750|M.tuberculosis_H37Rv ASV----PYPLSTADDLAATVELVEDAGGGIVARQGDVRDRASLSVALQA
MUL_3392|M.ulcerans_Agy99 ASV----PYPLGTAEELATTVKLVEDTGARIVASQADVRDREALAAALQA
MSMEG_4849|M.smegmatis_MC2_155 RPG-AETAIPASTSDDLAETANLVKGLDRRVVTAEVDVRDAAAIKAAVDS
MAV_0896|M.avium_104 RAGVVDTAIPASTPEDLAETADLVKGHNRRIVTAEVDVRDYDALKAAVDS
MMAR_4754|M.marinum_M VEN---TTIPASTPEDLAETADLVKGHNRRIFTAEADVRDYDALKAAVDA
TH_3237|M.thermoresistible__bu DSV----QIPLASPEDLAETADLVKGLNRRIYTAEVDVRDFDALKAAVDT
Mflv_1589|M.gilvum_PYR-GCK STH---TDIPPATPDDLAQTVDLVKGTGRRIVSEQVDVRDFAAVKAAVDS
Mvan_5168|M.vanbaalenii_PYR-1 STH---TDIPAATPDDLAQTVDLVKGIGRRVVAAEVDVRDYAAVKAAVDS
MAB_0072c|M.abscessus_ATCC_199 HTT----ITPPATRADLDKTSELVESAGRRVIPGVVDVRNLRDVQTFTQD
MLBr_01807|M.leprae_Br4923 VAD--------GHRVAVTHRASVVP---DWFFGVECDVTDNDAIGAAFKA
. : .:* . ** : : . .
Mb2771|M.bovis_AF2122/97 GLDEFGRLDIVVANAGIA-----MMQAGDDGWRDVIDVNLTGVFHTVQVA
Rv2750|M.tuberculosis_H37Rv GLDEFGRLDIVVANAGIA-----MMQAGDDGWRDVIDVNLTGVFHTVQVA
MUL_3392|M.ulcerans_Agy99 GIDELGQVDIVVANAGIA-----PMQSGDDGWRDVIDVNLSGAYYTVEVA
MSMEG_4849|M.smegmatis_MC2_155 GVEQLGRLDIIVANAGIGNGGDTLDQTSEEDWDEMIDINLSGVWKSVKAG
MAV_0896|M.avium_104 GVEQLGRLDIIVANAGIGNGGDTLDKTSEEDWTEMIDINLAGVWKTVKAG
MMAR_4754|M.marinum_M GVDELGRLDIIVANAGIGNGGATLDKTSEHDWQEMIDVNLSGVWKSVKAA
TH_3237|M.thermoresistible__bu GVEQLGRLDIIVANAGIGNGGETLDKTSEGDWTDMIDVNLAGVWKTVKAG
Mflv_1589|M.gilvum_PYR-GCK GVEQLGRLDIVVANAGIGNGGQTLDHTSEEDWNDMIDVNLSGVWKSVKAA
Mvan_5168|M.vanbaalenii_PYR-1 GVEQLGRLDIVVANAGIGNGGQTLDHTSEEDWNDMIDVNLSGVWKSVKAA
MAB_0072c|M.abscessus_ATCC_199 AVTEFGGLDIVCATAGITSRRMLLDMP-ESDWQTMLDVNLTGVWHTTRAA
MLBr_01807|M.leprae_Br4923 VEEHQGPVEVLVANAGITS-DMLIMRMSEEQFQKVIDVNLTGVFRVAKLA
. * :::: *.*** : : ::*:**:*.: .. .
Mb2771|M.bovis_AF2122/97 IPTLIEQGTGGSIVLISSAAGLVGIGSSDPGSLGYAAAKHGVVGLMRAYA
Rv2750|M.tuberculosis_H37Rv IPTLIEQGTGGSIVLISSAAGLVGIGSSDPGSLGYAAAKHGVVGLMRAYA
MUL_3392|M.ulcerans_Agy99 IPTMIEQGRGGSIVLISSAAGLVGISSADAGAIGYVASKHALVGLMRVYA
MSMEG_4849|M.smegmatis_MC2_155 VPHIVAGGRGGSIVLTSSVGGLKAYPHCG----NYVAAKHGVVGLMRGFA
MAV_0896|M.avium_104 VPHMIAGGRGGSIILTSSVGGLKAYPHTG----HYVAAKHGVVGLMRAFG
MMAR_4754|M.marinum_M VPHLIAGGNGGSIVLTSSVGGMKAYPHCG----NYVAAKHGVVGLMRSFA
TH_3237|M.thermoresistible__bu VPHILAGGRGGSIILTSSVGGLKAYPHTG----HYVAAKHGVVGLMRTFA
Mflv_1589|M.gilvum_PYR-GCK VPHLLAGGNGGSIILTSSVGGLKPYPHTG----HYIAAKHGVIGLMRTFA
Mvan_5168|M.vanbaalenii_PYR-1 VPHLLSGGRGGSIILTSSVGGLKPYAHTG----HYIAAKHGVIGLMRTFA
MAB_0072c|M.abscessus_ATCC_199 TPHLIARG-GGAMVLVSSIAGLRGLVGVS----HYVAAKHGVVGLMRSLA
MLBr_01807|M.leprae_Br4923 SRNMIKQRFGRYIFMG-SVVGLSGVPGQA----NYAASKSGLIGMARSLA
:: * :.: * *: * *:* .::*: * .
Mb2771|M.bovis_AF2122/97 NHLAPQNIRVNSVHPCGVDTPMINNEFFQQWLT-------TADMDAPHNL
Rv2750|M.tuberculosis_H37Rv NHLAPQNIRVNSVHPCGVDTPMINNEFFQQWLT-------TADMDAPHNL
MUL_3392|M.ulcerans_Agy99 NLLAPHSIRVNSLHPSGVDPPMINNEFIRHWLAD-----LVAETGSGPGA
MSMEG_4849|M.smegmatis_MC2_155 VELGQHNIRVNTVHPTHVATPMLHNEGTFKMFRPDLENPGPDDMAPICQM
MAV_0896|M.avium_104 VELGQHMIRVNSVHPTHVKTPMLHNEGTFKMFRPDLENPGPDDMAPICQM
MMAR_4754|M.marinum_M VELGQHMIRVNSVHPTHVRTPMLHNEGTFKMFRPDLENPGPDDMAPICQL
TH_3237|M.thermoresistible__bu VELGQHNIRVNSVHPTNVNTPLFMNEPTMKLFRPDLENPGPEDLKVVAQM
Mflv_1589|M.gilvum_PYR-GCK VELGQHSIRVNAVCPTNVNTPLFMNEGTMKLFRPDLENPGPDDLAVAAQF
Mvan_5168|M.vanbaalenii_PYR-1 VELGQHSIRVNAVCPTNVNTPLFMNEGTMKLFRPDLENPGPDDLAVAAQF
MAB_0072c|M.abscessus_ATCC_199 IELAPHKIRVNTVHPTNVDTPMIQNEFVRTLFRPDLEQPTREDFAAAAKT
MLBr_01807|M.leprae_Br4923 RELGSRSITANVVAPGFIDTEMTR----------------AMDSKYQEAA
*. : * .* : * : . : :
Mb2771|M.bovis_AF2122/97 GNALPVELV-QPTDIANAVAWLASEEARYVTGVTLPVDAGFVNKR
Rv2750|M.tuberculosis_H37Rv GNALPVELV-QPTDIANAVAWLASEEARYVTGVTLPVDAGFVNKR
MUL_3392|M.ulcerans_Agy99 GNALPVQIL-QADDIAGALAWLVSDEARYITGVALPVDAGCVNKR
MSMEG_4849|M.smegmatis_MC2_155 FHTLPIPWV-EPIDISNAVLFLASDEARYITGVTLPVDAGSCLK-
MAV_0896|M.avium_104 FHTLPIPWV-EPIDISNAVLFFASDEARYITGVTLPIDAGSCLK-
MMAR_4754|M.marinum_M FHTLPIPWV-EAEDISNAVLFLASDESRYITGVTLPVDAGGCLK-
TH_3237|M.thermoresistible__bu MHTLPIGWV-EPEDIANAVLFLASDEARYITGVTLPVDGGSCLK-
Mflv_1589|M.gilvum_PYR-GCK MHVLPVGWV-EPRDISNAVLFLASDEARYVTGLPVTVDAGSMLK-
Mvan_5168|M.vanbaalenii_PYR-1 MHVLPVGWV-EPVDISNAVLFLASDEARYITGLPVTVDAGSMLK-
MAB_0072c|M.abscessus_ATCC_199 MNLLDLPWV-EPLDIANASLFLASDEARYITSVSLPVDAGSTQR-
MLBr_01807|M.leprae_Br4923 LQAIPLGRIGAVEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH
: : : : :*:.* ::.*::: *::. :.:*.*