For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AGRVEGKVAFITGAARGQGRSHAVRLAEEGADIIAVDVCRRISSNEDIPASTPEDLAETVELVKGLNRRI VAEEVDVRDYDALKAVVDSGVEQLGGLDIVVANAGIGNGGATLDKTSEADWDDMIGVNLSGVWKTVKAAV PHLISGGNGGSIILTSSVGGLKAYPHTGHYIAAKHGVVGLMRTFAVELGQHSIRVNSVHPTNVNTPLFMN EGTMKLFRPDLENPGPDDMAVVAQMMHVLPVGWVEPRDISNAVLFLASDEARYVTGLPMTVDAGSMLK*
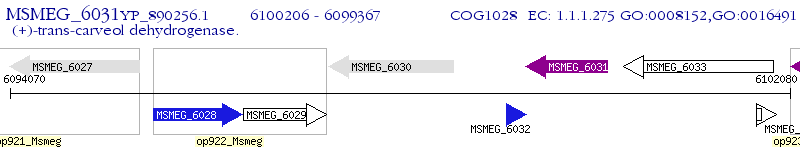
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6031 | - | - | 100% (279) | carveol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_4849 | - | e-123 | 75.44% (281) | carveol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1410 | fabG | 2e-68 | 47.69% (281) | 3-ketoacyl-(acyl-carrier-protein) reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0706 | fabG | 1e-67 | 47.64% (275) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_1589 | - | 1e-139 | 86.74% (279) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0687 | fabG | 1e-67 | 47.64% (275) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01740 | - | 6e-23 | 30.91% (220) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0072c | - | 5e-78 | 53.62% (276) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_4754 | - | 1e-127 | 78.14% (279) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_0846 | - | 1e-138 | 86.02% (279) | carveol dehydrogenase |
| M. thermoresistible (build 8) | TH_3237 | - | 1e-134 | 82.08% (279) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3392 | fabG | 8e-61 | 46.98% (281) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_5168 | - | 1e-138 | 86.38% (279) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0706|M.bovis_AF2122/97 -MSARGGSLHGRVAFVTGAARAQGRSHAVRLAREGADIVALDICAPVSGS
Rv0687|M.tuberculosis_H37Rv -MSARGGSLHGRVAFVTGAARAQGRSHAVRLAREGADIVALDICAPVSGS
Mflv_1589|M.gilvum_PYR-GCK ----MGGRVEGKVAFITGAARGQGRSHAVRLAEEGADIIAIDVCGPISTH
Mvan_5168|M.vanbaalenii_PYR-1 ----MGGRVEGKVAFITGAARGQGRSHAVRLAEEGADIIAIDVCGPISTH
MSMEG_6031|M.smegmatis_MC2_155 ----MAGRVEGKVAFITGAARGQGRSHAVRLAEEGADIIAVDVCRRISSN
MAV_0846|M.avium_104 ----MGGRVEGKVAFITGAARGQGRSHAVRLAQEGADIIAVDICAPISSN
TH_3237|M.thermoresistible__bu ----MAGRLEGKVAFITGAARGQGRAHAVRMAQEGADIIAVDICRQIDS-
MMAR_4754|M.marinum_M MTEQLGGRVAGKVAFITGAARGQGRSHAVRLAQEGADIIAVDVCKPIVEN
MAB_0072c|M.abscessus_ATCC_199 ----MTGRLTGKVALVTGAARGIGRAQAVRFAQEGADIIALDICAPVHT-
MUL_3392|M.ulcerans_Agy99 MSRPLDGPLSGRVAFITGAARGQGRAHALRLARDGADVIAVDLCDQIAS-
MLBr_01740|M.leprae_Br4923 -MVKTAQYFVGKRCFVTGAASGIGRATALRLAAYGAELYLTDR-------
. *: .::**** . **: *:*:* **:: *
Mb0706|M.bovis_AF2122/97 VTYPPATSEDLGETVRAVEAEGRKVLA-REVDIRDDAELRRLVADGVEQF
Rv0687|M.tuberculosis_H37Rv VTYPPATSEDLGETVRAVEAEGRKVLA-REVDIRDDAELRRLVADGVEQF
Mflv_1589|M.gilvum_PYR-GCK TDIPPATPDDLAQTVDLVKGTGRRIVS-EQVDVRDFAAVKAAVDSGVEQL
Mvan_5168|M.vanbaalenii_PYR-1 TDIPAATPDDLAQTVDLVKGIGRRVVA-AEVDVRDYAAVKAAVDSGVEQL
MSMEG_6031|M.smegmatis_MC2_155 EDIPASTPEDLAETVELVKGLNRRIVA-EEVDVRDYDALKAVVDSGVEQL
MAV_0846|M.avium_104 SQIPPSTPDDLAETADLIKGLDRRIVT-AEVDVRDYDALKAAVDSGVEQL
TH_3237|M.thermoresistible__bu VQIPLASPEDLAETADLVKGLNRRIYT-AEVDVRDFDALKAAVDTGVEQL
MMAR_4754|M.marinum_M TTIPASTPEDLAETADLVKGHNRRIFT-AEADVRDYDALKAAVDAGVDEL
MAB_0072c|M.abscessus_ATCC_199 TITPPATRADLDKTSELVESAGRRVIP-GVVDVRNLRDVQTFTQDAVTEF
MUL_3392|M.ulcerans_Agy99 VPYPLGTAEELATTVKLVEDTGARIVA-SQADVRDREALAAALQAGIDEL
MLBr_01740|M.leprae_Br4923 ------HCDGLAQTVSEARALGAQVPEYRALDISDYDEVAAFGADIHARH
* * . . :: *: : : .
Mb0706|M.bovis_AF2122/97 GRLDIVVANAGVLGWGRLWELT-DEQWETVIGVNLTGTWRTLRATVPAMI
Rv0687|M.tuberculosis_H37Rv GRLDIVVANAGVLGWGRLWELT-DEQWETVIGVNLTGTWRTLRATVPAMI
Mflv_1589|M.gilvum_PYR-GCK GRLDIVVANAGIGNGGQTLDHTSEEDWNDMIDVNLSGVWKSVKAAVPHLL
Mvan_5168|M.vanbaalenii_PYR-1 GRLDIVVANAGIGNGGQTLDHTSEEDWNDMIDVNLSGVWKSVKAAVPHLL
MSMEG_6031|M.smegmatis_MC2_155 GGLDIVVANAGIGNGGATLDKTSEADWDDMIGVNLSGVWKTVKAAVPHLI
MAV_0846|M.avium_104 GRLDIICANAGIGNGGQTLDKTSEDDWRDMIDVNLSGVWKTVKAGVPHLI
TH_3237|M.thermoresistible__bu GRLDIIVANAGIGNGGETLDKTSEGDWTDMIDVNLAGVWKTVKAGVPHIL
MMAR_4754|M.marinum_M GRLDIIVANAGIGNGGATLDKTSEHDWQEMIDVNLSGVWKSVKAAVPHLI
MAB_0072c|M.abscessus_ATCC_199 GGLDIVCATAGITSRRMLLDMP-ESDWQTMLDVNLTGVWHTTRAATPHLI
MUL_3392|M.ulcerans_Agy99 GQVDIVVANAGIAP-----MQSGDDGWRDVIDVNLSGAYYTVEVAIPTMI
MLBr_01740|M.leprae_Br4923 PSMDIVMNIAGVSVWG-TVDRLTHDQWSNVVAINLMGPIHIIETFVPAIL
:**: **: . * :: :** * .. * ::
Mb0706|M.bovis_AF2122/97 DAGNGGSIVVVSSSAGLKATPGNGH----YAASKHALVALTNTLAIELGE
Rv0687|M.tuberculosis_H37Rv DAGNGGSIVVVSSSAGLKATPGNGH----YAASKHALVALTNTLAIELGE
Mflv_1589|M.gilvum_PYR-GCK AGGNGGSIILTSSVGGLKPYPHTGH----YIAAKHGVIGLMRTFAVELGQ
Mvan_5168|M.vanbaalenii_PYR-1 SGGRGGSIILTSSVGGLKPYAHTGH----YIAAKHGVIGLMRTFAVELGQ
MSMEG_6031|M.smegmatis_MC2_155 SGGNGGSIILTSSVGGLKAYPHTGH----YIAAKHGVVGLMRTFAVELGQ
MAV_0846|M.avium_104 SQGQGGSVILTSSVGGLKAYPHTGH----YIAAKHGVVGLMRTFAVELGQ
TH_3237|M.thermoresistible__bu AGGRGGSIILTSSVGGLKAYPHTGH----YVAAKHGVVGLMRTFAVELGQ
MMAR_4754|M.marinum_M AGGNGGSIVLTSSVGGMKAYPHCGN----YVAAKHGVVGLMRSFAVELGQ
MAB_0072c|M.abscessus_ATCC_199 ARG-GGAMVLVSSIAGLRGLVGVSH----YVAAKHGVVGLMRSLAIELAP
MUL_3392|M.ulcerans_Agy99 EQGRGGSIVLISSAAGLVGISSADAGAIGYVASKHALVGLMRVYANLLAP
MLBr_01740|M.leprae_Br4923 AAGRGGHLVNVSSAAGLVALPWHAA----YSASKYGLRGLSEVLRFDLAR
* ** :: ** .*: * *:*:.: .* . *.
Mb0706|M.bovis_AF2122/97 FGIRVNSIHPYSVDTPMIEPEAMIQTFAKHPG------YVHSFPPMPLQP
Rv0687|M.tuberculosis_H37Rv FGIRVNSIHPYSVDTPMIEPEAMIQTFAKHPG------YVHSFPPMPLQP
Mflv_1589|M.gilvum_PYR-GCK HSIRVNAVCPTNVNTPLFMNEGTMKLFRPDLENPGPDDLAVAAQFMHVLP
Mvan_5168|M.vanbaalenii_PYR-1 HSIRVNAVCPTNVNTPLFMNEGTMKLFRPDLENPGPDDLAVAAQFMHVLP
MSMEG_6031|M.smegmatis_MC2_155 HSIRVNSVHPTNVNTPLFMNEGTMKLFRPDLENPGPDDMAVVAQMMHVLP
MAV_0846|M.avium_104 HFIRVNSVHPTNVNTPMFMNEGTMRLFRPDLKNPGPDDLEVAAQFMHVLP
TH_3237|M.thermoresistible__bu HNIRVNSVHPTNVNTPLFMNEPTMKLFRPDLENPGPEDLKVVAQMMHTLP
MMAR_4754|M.marinum_M HMIRVNSVHPTHVRTPMLHNEGTFKMFRPDLENPGPDDMAPICQLFHTLP
MAB_0072c|M.abscessus_ATCC_199 HKIRVNTVHPTNVDTPMIQNEFVRTLFRPDLEQPTREDFAAAAKTMNLLD
MUL_3392|M.ulcerans_Agy99 HSIRVNSLHPSGVDPPMINNEFIRHWLADLVAETGSG-----PGAGNALP
MLBr_01740|M.leprae_Br4923 YRIGVSVVVPGAVRTPLVDTIEIAGVDRQDPRFSRWVDRFRGHAVSAERA
. * *. : * * .*:.
Mb0706|M.bovis_AF2122/97 KGFMTPDEISDVVVWLAGDGSGALSGNQ----------------------
Rv0687|M.tuberculosis_H37Rv KGFMTPDEISDVVVWLAGDGSGALSGNQ----------------------
Mflv_1589|M.gilvum_PYR-GCK VGWVEPRDISNAVLFLASDEARYVTGLP----------------------
Mvan_5168|M.vanbaalenii_PYR-1 VGWVEPVDISNAVLFLASDEARYITGLP----------------------
MSMEG_6031|M.smegmatis_MC2_155 VGWVEPRDISNAVLFLASDEARYVTGLP----------------------
MAV_0846|M.avium_104 VGWVEPVDISNAVLFLASDESRYITGLP----------------------
TH_3237|M.thermoresistible__bu IGWVEPEDIANAVLFLASDEARYITGVT----------------------
MMAR_4754|M.marinum_M IPWVEAEDISNAVLFLASDESRYITGVT----------------------
MAB_0072c|M.abscessus_ATCC_199 LPWVEPLDIANASLFLASDEARYITSVS----------------------
MUL_3392|M.ulcerans_Agy99 VQILQADDIAGALAWLVSDEARYITGVA----------------------
MLBr_01740|M.leprae_Br4923 AEKILAGVAKNRYLIYTSHDIRVLYGFKRLAWWPYGVVMKQVNVVFTRAL
: . . ... : .
Mb0706|M.bovis_AF2122/97 ----------IPVDKGALKY-----------
Rv0687|M.tuberculosis_H37Rv ----------IPVDKGALKY-----------
Mflv_1589|M.gilvum_PYR-GCK ----------VTVDAGSMLK-----------
Mvan_5168|M.vanbaalenii_PYR-1 ----------VTVDAGSMLK-----------
MSMEG_6031|M.smegmatis_MC2_155 ----------MTVDAGSMLK-----------
MAV_0846|M.avium_104 ----------VTVDAGSMLK-----------
TH_3237|M.thermoresistible__bu ----------LPVDGGSCLK-----------
MMAR_4754|M.marinum_M ----------LPVDAGGCLK-----------
MAB_0072c|M.abscessus_ATCC_199 ----------LPVDAGSTQR-----------
MUL_3392|M.ulcerans_Agy99 ----------LPVDAGCVNKR----------
MLBr_01740|M.leprae_Br4923 RPSPATMRCVEPIKVGVYSQPRSGEGGKSGN
.:. *