For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KAVTVSDRDAGVSGLSLTELPYPVVSENDVVVRVHAAGFTRGELDWPDTWTERSGRDRTPSVPGHELSGV VVELGYGTTRFTIGQRVFGLADWARNGSLAEYAAVETRHLAPLPYDVDHTLAAALPISGLSAWQGLFEHA QLVAGQTVLIHGAAGGVGSIAVQLAREVGARVIGTGRAADRDRALELGTHTFLDLDDDKLEDAGEVDVVF DVIGGEIMERSIALVRAGGTLVTLAEPPKTQPRDGRALYFLVEPDSARLADLAARVRTGRLKPVVGRVVP LADAPSAFNQKKRTTGKTIVRVTED*
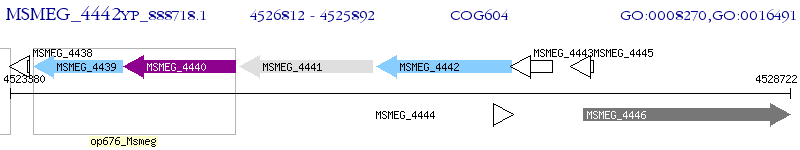
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4442 | - | - | 100% (306) | zinc-binding oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_5762 | - | e-115 | 64.38% (306) | zinc-binding oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_4828 | - | 9e-25 | 29.58% (311) | alcohol dehydrogenase, zinc-binding |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3806 | - | 5e-24 | 26.61% (342) | putative oxidoreductase |
| M. gilvum PYR-GCK | Mflv_2124 | - | 4e-35 | 33.33% (297) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv3777 | - | 1e-23 | 26.32% (342) | oxidoreductase |
| M. leprae Br4923 | MLBr_00118 | - | 6e-22 | 27.88% (226) | putative oxidireductase |
| M. abscessus ATCC 19977 | MAB_3307c | - | 1e-111 | 63.37% (303) | putative zinc-binding oxidoreductase |
| M. marinum M | MMAR_1738 | - | 2e-59 | 41.72% (302) | oxidoreductase |
| M. avium 104 | MAV_0242 | - | 7e-28 | 29.46% (336) | quinone oxidoreductase |
| M. thermoresistible (build 8) | TH_1665 | - | 4e-26 | 34.68% (248) | PROBABLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_2209 | - | 5e-26 | 32.15% (311) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_5615 | - | 3e-26 | 28.66% (321) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00118|M.leprae_Br4923 ------MHAIVAESAN-QMVWREVPDVTAGPGEVLIEVAASGVNR-----
MAV_0242|M.avium_104 ---MATMRAIVAESAD-QLSWQEVADVSAGPGEVVVKVAAAGVNR-----
Mb3806|M.bovis_AF2122/97 ---MTIMRAVVAESSD-RLVWQEVPDVSAGPGEVLIKVAASGVNR-----
Rv3777|M.tuberculosis_H37Rv ---MTIMRAVVAESSD-RLVWQEVPDVSAGPGEVLIKVAASGVNR-----
TH_1665|M.thermoresistible__bu ------MRAISVQTSG-QLSWVEVPDIEPGESEVLIAVHTAGINR-----
Mvan_5615|M.vanbaalenii_PYR-1 ------MHAIVAQPSSEGLTWQSVPDIAPAPGEVLIRVAAAGVNR-----
MSMEG_4442|M.smegmatis_MC2_155 ---MKAVTVSDRDAGVSGLSLTELPYPVVSENDVVVRVHAAGFTRG----
MAB_3307c|M.abscessus_ATCC_199 ---MKAI-VAEQGAKGAGLSLTAVPYPHAAENDVIVQVHAAGFTPG----
MMAR_1738|M.marinum_M ---MRALRAHRRGGPET-LAVERAPVPVPAAGEVVVAVRAAGITFD----
Mflv_2124|M.gilvum_PYR-GCK MSDRMKAVVLARFGGADAFELRDVAVPHVGPRQVRVRVHATAVNP-----
MUL_2209|M.ulcerans_Agy99 ----MKAMAQRTWNPAEPLEFIELPTPEPQAGEVRVQVQAIGVNPVDWKM
. : . :* : * : ...
MLBr_00118|M.leprae_Br4923 -ADVLQVAGKYPHPPGASAIIGMEVAGVVAAVSSGVTEWSVGQEVCALLA
MAV_0242|M.avium_104 -ADVLQAAGKYPPPPGASETIGMEVSGVIAEVGDGVTEWSVGQEVCALLA
Mb3806|M.bovis_AF2122/97 -ADVLQAAGKYPPPPGVSDIIGLEVSGIVAAVGPGVTEWSAGQEVCALLA
Rv3777|M.tuberculosis_H37Rv -ADVLQAAGKYPPPPGVSDIIGLEVSGIVAAVGPGVTEWSAGQEVCALLA
TH_1665|M.thermoresistible__bu -ADLLQAAGKYPPPPGASEILGLEVSGTVAAVGAGVSGWTVGQPVCALLA
Mvan_5615|M.vanbaalenii_PYR-1 -ADLLQAAGKYPPPPGASDILGLEVSGTIAAIGEGVTKWSQGQPVCALLA
MSMEG_4442|M.smegmatis_MC2_155 ELDWPDTWTERSGRDRTPSVPGHELSGVVVELGYGTTRFTIGQRVFGLAD
MAB_3307c|M.abscessus_ATCC_199 ELDWPGTWADRSGRSRGPSIPGHEVSGIVVELGYGTTGLTVGQRVFGITD
MMAR_1738|M.marinum_M ELTWDQTWT-RDGVSRTPVIPSHEVSGVIHEIGSGTEDFHCGDEVYGLIG
Mflv_2124|M.gilvum_PYR-GCK -LDYQIRRGDYTDLVPLPAIIGHDISGVIEEVGSHVTEFRAGDAVYYTPK
MUL_2209|M.ulcerans_Agy99 RSSGPLRLAARLIGPKPPVVVGVDFAGVVEAVGPGVTRAAPGDRVVGGTN
. . :.:* : :. . *: *
MLBr_00118|M.leprae_Br4923 G-----GGYAEYVAVPASQVMPIPKGVNVVDSAGIPEVACTVWSNLVMAA
MAV_0242|M.avium_104 G-----GGYAEYVAVPAGQLLPIPAGVDLVDAAGLPEVACTVWSNLVLIA
Mb3806|M.bovis_AF2122/97 G-----GGYAEYVAVPADQVLPIPPSVNLVDSAALPEVACTVWSNLVMTA
Rv3777|M.tuberculosis_H37Rv G-----GGYAEYVAVPADQVLPIPPSVNLVDSAALPEVACTVWSNLVMTA
TH_1665|M.thermoresistible__bu G-----GGYAEYVAVPAEQVMPIPESVEIRYAAALPEVACTVWSNLVMTA
Mvan_5615|M.vanbaalenii_PYR-1 G-----GGYAEFVAVPAGQVLPIPDGVPVPHAAGLPEVACTVWSNVVMAA
MSMEG_4442|M.smegmatis_MC2_155 WARN--GSLAEYAAVETRHLAPLPYDVDHTLAAALPISGLSAWQGLFEHA
MAB_3307c|M.abscessus_ATCC_199 WTRN--GALAQYVAVEARNLAPLSADVDHTVAAALPVSGLTAWQGLFLHG
MMAR_1738|M.marinum_M FDRD--GAAADFVAVPAAHLAAKPATISHTVAAALPLAGLTALQALVDHA
Mflv_2124|M.gilvum_PYR-GCK IFGGP-GSYAEQHVADVDLVGRKPDNLDHLEAASLTLVGGTVWESLVTRA
MUL_2209|M.ulcerans_Agy99 FSRGQRGSYADTVLVREDQLCRVPDTVDIAVAAALPVGGVTAWMAVVELG
*. *: . : . : :*.:. . :. :. .
MLBr_00118|M.leprae_Br4923 HLS----AGQLLLLHGGTSGIGSHGIQVARALGAKVAVTAGSEEKLEFCR
MAV_0242|M.avium_104 GLR----NGQLLLVHGGASGIGSHAIQVARALGARVAVTAGSAEKLGFCR
Mb3806|M.bovis_AF2122/97 HLR----PGQLVLIHGGASGIGSHAIQVARALAARVAITAGSPEKLELCR
Rv3777|M.tuberculosis_H37Rv HLR----PGQLVLIHGGASGIGSHAIQVVRALAARVAITAGSPEKLELCR
TH_1665|M.thermoresistible__bu GLS----AGEWVLIHGGASGVGSHAVQVARALDARVAVTAGTRHKLELCR
Mvan_5615|M.vanbaalenii_PYR-1 GLT----APELLLVHGGASGIGTHAIQVAKALNCRVAVTAGSRNKLDLCA
MSMEG_4442|M.smegmatis_MC2_155 QLV----AGQTVLIHGAAGGVGSIAVQLAREVGARVIGTGRAAD-RDRAL
MAB_3307c|M.abscessus_ATCC_199 HIV----AGQSVLIHGAAGAVGSVAVQLAHEAGAHVIGTGRGGQ-RDQVL
MMAR_1738|M.marinum_M AVR----PGEAVLVHGGAGGVGALVVQLAAILGANVTATVRSDA-GELVS
Mflv_2124|M.gilvum_PYR-GCK QLT----VGETILIHGGAGGVGTIAIQVAKAMGARVITTAKAGD-HAFVN
MUL_2209|M.ulcerans_Agy99 RIRQVPSADRRVLVLGASGGVGQLAVQIAKLQEAFVVGVCSTEN-VEMVK
: . :*: *.:..:* :*:. . * .
MLBr_00118|M.leprae_Br4923 ALGAQITINYRDEDFVARLQQQN--GGADVILDIMGAAYLDRNIDALAID
MAV_0242|M.avium_104 ELGADITINYRDEDFVARLADET--GGADVILDIMGAAYLDRNIDALATD
Mb3806|M.bovis_AF2122/97 DLGAQITINYRDEDFVARLKQETDGSGADIILDIMGASYLDRNIDALATD
Rv3777|M.tuberculosis_H37Rv DLGAQITINYRDEDFVARLKQETDGSGADIILDIMGASYLDRNIDALATD
TH_1665|M.thermoresistible__bu ELGAEVTICYREEDFVERVRAETDGAGADVVLDIMGAAYLGRNVDALATD
Mvan_5615|M.vanbaalenii_PYR-1 ELGADITIDYHNEDYVGVVR---DAGGADVILDIMGAAYLDRNIETLADG
MSMEG_4442|M.smegmatis_MC2_155 ELGTHTFLDLDDDKLEDAG-------EVDVVFDVIGGEIMERSIALVRAG
MAB_3307c|M.abscessus_ATCC_199 GLKADDFVDLTTDTLEDIG-------QVDVVFDVIGGEILDRSAQLVRAG
MMAR_1738|M.marinum_M RLGAQRVIDTRTEAFERTAT------DFDVVIDTVGGDTLDRSFAVLRPG
Mflv_2124|M.gilvum_PYR-GCK SLGADVAIDHTSTDYVDAVAEATRGRGVDVVFDTIGGDTLTRSPLSLADS
MUL_2209|M.ulcerans_Agy99 DLGADLVLDYNQGDALAQAKP---HAPFHVVIDCVGSYSGAGCRALLSSR
* :. : .:::* :*. :
MLBr_00118|M.leprae_Br4923 GQLIVIGLQGGVKGELNLGKVLSKRARIIGSTLRARPVNGPNSKSEIVAA
MAV_0242|M.avium_104 GQLVIIGMQGGVKAELNIGKLLAKRARVIGTTLRGRPVSGPNSKTEIVQA
Mb3806|M.bovis_AF2122/97 GQLIVIGMQGGVKAELNLGKLLTKRARVIGTTLRARPVSGPHGKAAIAQA
Rv3777|M.tuberculosis_H37Rv GQLIVIGMQGGVKAELNLGKLLTKRARVIGTTLRARPVSGPHGKAAIAQA
TH_1665|M.thermoresistible__bu GRLVVIGMQGGRRAELDLGKLVAKRAAVFGTALRGRPVTGPHGKADIVRA
Mvan_5615|M.vanbaalenii_PYR-1 GRLVIIGMQGGAKAELNIGKLLTKRGSVIATALRSRPVHGRGGKADIVAR
MSMEG_4442|M.smegmatis_MC2_155 GTLVTLAEPP---------KTQPRDGRALYFLVEPD------------SA
MAB_3307c|M.abscessus_ATCC_199 GALITIAEPP---------RVIPEGARAVFFVVEAD------------RR
MMAR_1738|M.marinum_M GRLVTLSAPPPA------GKADEHGVTAMFFIVTPN------------RD
Mflv_2124|M.gilvum_PYR-GCK GRVVSIVDIARPQ---NLIEAWGRNAAYHFVFTRQN------------RG
MUL_2209|M.ulcerans_Agy99 GRHIMVAGDEPSS----AVQVVVPPFKSKAILGRPN------------GA
* : : .
MLBr_00118|M.leprae_Br4923 VTASIWPMIADGSVRPIIGARMAVQQAADAHQLLLSG---KVSGKIVLTV
MAV_0242|M.avium_104 VTASVWPMIADGRVRPIIGARLPIQQAGDAHRRLVAS---EVTGKIVLTV
Mb3806|M.bovis_AF2122/97 VAASVWPMIAANRVRPVIGTRLPIQQAAQAHELMLSG---KTFGKILLTV
Rv3777|M.tuberculosis_H37Rv VAASVWPMIAANRVRPVIGTRLPIQQAAQAHELMLSG---KTFGKILLTV
TH_1665|M.thermoresistible__bu VVENVWPMVASDQVRPVVGAEFPIQEAQAAHELLASG---DVYGKVLLCV
Mvan_5615|M.vanbaalenii_PYR-1 VGAELWPLVADGQVRPVIGAEFPIAEAQAAHELLASG---DVSGKVLLRV
MSMEG_4442|M.smegmatis_MC2_155 RLADLAARVRTGRLKPVVGRVVPLADAPSAFNQKK-----RTTGKTIVRV
MAB_3307c|M.abscessus_ATCC_199 QLTILENKLRDGRIRQNIGSVIPLEDALSAFDIDR-----KTPGKTIVQI
MMAR_1738|M.marinum_M QLNRLGELVGSGRLHVEIAQSFPLANGREAFESGRGAA--RRAGKTVITV
Mflv_2124|M.gilvum_PYR-GCK KLDALTMLVERGLVKPVLGATLPLARMGEAHELLENRRSYALRGKVAIDV
MUL_2209|M.ulcerans_Agy99 RLEPLVDAVAAGKLRVSIAQKLPLTSAEEAHRLSQTS---RMTGKLILEP
: : . :: :. ..: *. ** :
MLBr_00118|M.leprae_Br4923 AGSGTETLLSRHH
MAV_0242|M.avium_104 -------------
Mb3806|M.bovis_AF2122/97 -------------
Rv3777|M.tuberculosis_H37Rv -------------
TH_1665|M.thermoresistible__bu ED-----------
Mvan_5615|M.vanbaalenii_PYR-1 DV-----------
MSMEG_4442|M.smegmatis_MC2_155 TED----------
MAB_3307c|M.abscessus_ATCC_199 AAV----------
MMAR_1738|M.marinum_M GQ-----------
Mflv_2124|M.gilvum_PYR-GCK AGETVALPPRIS-
MUL_2209|M.ulcerans_Agy99 -------------