For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDRMKAVVLARFGGADAFELRDVAVPHVGPRQVRVRVHATAVNPLDYQIRRGDYTDLVPLPAIIGHDISG VIEEVGSHVTEFRAGDAVYYTPKIFGGPGSYAEQHVADVDLVGRKPDNLDHLEAASLTLVGGTVWESLVT RAQLTVGETILIHGGAGGVGTIAIQVAKAMGARVITTAKAGDHAFVNSLGADVAIDHTSTDYVDAVAEAT RGRGVDVVFDTIGGDTLTRSPLSLADSGRVVSIVDIARPQNLIEAWGRNAAYHFVFTRQNRGKLDALTML VERGLVKPVLGATLPLARMGEAHELLENRRSYALRGKVAIDVAGETVALPPRIS*
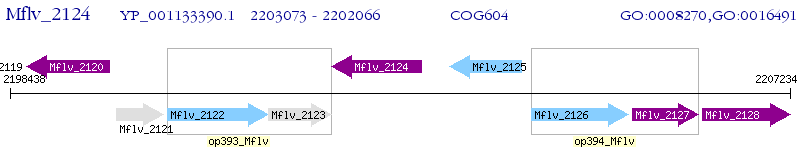
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2124 | - | - | 100% (335) | alcohol dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1189 | - | 2e-32 | 34.57% (324) | alcohol dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3686 | - | 3e-30 | 31.60% (326) | alcohol dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3806 | - | 8e-34 | 31.04% (335) | putative oxidoreductase |
| M. tuberculosis H37Rv | Rv3777 | - | 2e-33 | 30.75% (335) | oxidoreductase |
| M. leprae Br4923 | MLBr_00118 | - | 3e-30 | 30.00% (320) | putative oxidireductase |
| M. abscessus ATCC 19977 | MAB_0207c | - | 1e-39 | 34.21% (342) | quinone oxidoreductase |
| M. marinum M | MMAR_5332 | - | 6e-33 | 33.65% (318) | oxidoreductase |
| M. avium 104 | MAV_0242 | - | 1e-34 | 33.43% (338) | quinone oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_5762 | - | 6e-36 | 33.74% (326) | zinc-binding oxidoreductase |
| M. thermoresistible (build 8) | TH_1665 | - | 4e-32 | 32.84% (335) | PROBABLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_0096 | - | 4e-33 | 33.65% (318) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_5758 | - | 2e-44 | 40.00% (310) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00118|M.leprae_Br4923 ----MHAIVAESA---NQMVWREVPDVTAGPGEVLIEVAASGVNR--ADV
MAV_0242|M.avium_104 -MATMRAIVAESA---DQLSWQEVADVSAGPGEVVVKVAAAGVNR--ADV
Mb3806|M.bovis_AF2122/97 -MTIMRAVVAESS---DRLVWQEVPDVSAGPGEVLIKVAASGVNR--ADV
Rv3777|M.tuberculosis_H37Rv -MTIMRAVVAESS---DRLVWQEVPDVSAGPGEVLIKVAASGVNR--ADV
MMAR_5332|M.marinum_M -MAFVHAIVAESA---DHLVWQQVPDVTAGAGEVVIKVAAAGVNR--ADV
MUL_0096|M.ulcerans_Agy99 -MAFVHAIVAESA---DHLVWQQVPDVTAGAGEVVIKVAAAGVNR--ADV
TH_1665|M.thermoresistible__bu ----MRAISVQTS---GQLSWVEVPDIEPGESEVLIAVHTAGINR--ADL
MAB_0207c|M.abscessus_ATCC_199 ----MHAITVDNPGGPEALRWTEVPDPVPGPGEILIDVTAAAANR--ADL
Mflv_2124|M.gilvum_PYR-GCK MSDRMKAVVLARFGGADAFELRDVAVPHVGPRQVRVRVHATAVNP--LDY
MSMEG_5762|M.smegmatis_MC2_155 ---MRAIIAKDRTAGIDGLALAEVPYPHAAENDVIVAVHAAAFTPGELDW
Mvan_5758|M.vanbaalenii_PYR-1 ---MRAAVLTAYNS---PLLVDELPDPEPGPGEVLVRVMASGVNP--LDI
: : ::. . :: : * ::. . *
MLBr_00118|M.leprae_Br4923 LQVAGKYPHPPG-ASAIIGMEVAGVVAAVSSGVTEWSVGQEVCALLA---
MAV_0242|M.avium_104 LQAAGKYPPPPG-ASETIGMEVSGVIAEVGDGVTEWSVGQEVCALLA---
Mb3806|M.bovis_AF2122/97 LQAAGKYPPPPG-VSDIIGLEVSGIVAAVGPGVTEWSAGQEVCALLA---
Rv3777|M.tuberculosis_H37Rv LQAAGKYPPPPG-VSDIIGLEVSGIVAAVGPGVTEWSAGQEVCALLA---
MMAR_5332|M.marinum_M LQAAGKYPPPPG-ASEIIGMEVSGTITDIGDGVTEWSAGQEVCALLA---
MUL_0096|M.ulcerans_Agy99 LQAAGKYPPPPG-ASEIIGMEVSGTITDIGDGVTEWSAGQEVCALLA---
TH_1665|M.thermoresistible__bu LQAAGKYPPPPG-ASEILGLEVSGTVAAVGAGVSGWTVGQPVCALLA---
MAB_0207c|M.abscessus_ATCC_199 LQRRGLYSPPPG-ASSILGLECSGTVAALGADVAHWSVGQPVCALLS---
Mflv_2124|M.gilvum_PYR-GCK QIRRGDY-TDLVPLPAIIGHDISGVIEEVGSHVTEFRAGDAVYYTPKIFG
MSMEG_5762|M.smegmatis_MC2_155 PGTWTDR-AGRDRTPSVPGHEVSGVVAELGFGTTGLAVGQRVFGLTDWCR
Mvan_5758|M.vanbaalenii_PYR-1 KIRRGEAPHAQLPPPAVLGIDMAGVVEAVGPRVDRFTVGDEVFGMTGGVG
. * : :* : :. . .*: *
MLBr_00118|M.leprae_Br4923 --GGGYAEYVAVPASQVMPIPKGVNVVDSAGIPEVACTVWSNLVMAAHLS
MAV_0242|M.avium_104 --GGGYAEYVAVPAGQLLPIPAGVDLVDAAGLPEVACTVWSNLVLIAGLR
Mb3806|M.bovis_AF2122/97 --GGGYAEYVAVPADQVLPIPPSVNLVDSAALPEVACTVWSNLVMTAHLR
Rv3777|M.tuberculosis_H37Rv --GGGYAEYVAVPADQVLPIPPSVNLVDSAALPEVACTVWSNLVMTAHLR
MMAR_5332|M.marinum_M --GGGYAEYVAVPAGQVLPMPPGVSLIDAAALPEVACTVWSNLVMTAHLG
MUL_0096|M.ulcerans_Agy99 --GGGYAEYVAVPAGQVLPMPPGVSLIDAAALPEVACTVWSNLVMTAHLG
TH_1665|M.thermoresistible__bu --GGGYAEYVAVPAEQVMPIPESVEIRYAAALPEVACTVWSNLVMTAGLS
MAB_0207c|M.abscessus_ATCC_199 --GGGYAEKVVAPATQLLPVPTNVQLDVAAALPEVACTVWSNMMMTTKMS
Mflv_2124|M.gilvum_PYR-GCK GP-GSYAEQHVADVDLVGRKPDNLDHLEAASLTLVGGTVWESLVTRAQLT
MSMEG_5762|M.smegmatis_MC2_155 N--GTLAEYVAVEARNLAPLSATIDHTTAAALPISGLTAWQGLFTHGHLT
Mvan_5758|M.vanbaalenii_PYR-1 GLQGSLAELAAVDARLIAHKPRTLSMAQAAALPLGFITSWEGLVDRAGVG
* ** .. . : . :. :*.:. * *..:. :
MLBr_00118|M.leprae_Br4923 AGQLLLLHGGTSGIGSHGIQVARALGAKVAVTAGSEEKLEFCRALGAQIT
MAV_0242|M.avium_104 NGQLLLVHGGASGIGSHAIQVARALGARVAVTAGSAEKLGFCRELGADIT
Mb3806|M.bovis_AF2122/97 PGQLVLIHGGASGIGSHAIQVARALAARVAITAGSPEKLELCRDLGAQIT
Rv3777|M.tuberculosis_H37Rv PGQLVLIHGGASGIGSHAIQVVRALAARVAITAGSPEKLELCRDLGAQIT
MMAR_5332|M.marinum_M PGQLVLIHGGASGIGSHAIQVCTALGARVAVTAGSSEKLQLCRDLGAQIT
MUL_0096|M.ulcerans_Agy99 PGQLVLIHGGACGIGSHAIQVCTALGARVAVTAGSSEKLQLSRDLGAQIT
TH_1665|M.thermoresistible__bu AGEWVLIHGGASGVGSHAVQVARALDARVAVTAGTRHKLELCRELGAEVT
MAB_0207c|M.abscessus_ATCC_199 AGQSILIHGGASGIGTHTIQVAKALGLRVAVTAGTPEKLDACRDLGADLA
Mflv_2124|M.gilvum_PYR-GCK VGETILIHGGAGGVGTIAIQVAKAMGARVITTA-KAGDHAFVNSLGADVA
MSMEG_5762|M.smegmatis_MC2_155 PGETVLIHGAAGGVGSIAVQLAREAGAHVIGSG-RTADREYVLEVGAEAF
Mvan_5758|M.vanbaalenii_PYR-1 TGQQVLIHGGAGGVGFLAVQLALARGAQVYATG-STSSQAVIRAAG-AVP
*: :*:**.: *:* :*: :* :. . *
MLBr_00118|M.leprae_Br4923 INYRDEDFVARLQQQN--GGADVILDIMGAAYLDRNIDALAI-DGQLIVI
MAV_0242|M.avium_104 INYRDEDFVARLADET--GGADVILDIMGAAYLDRNIDALAT-DGQLVII
Mb3806|M.bovis_AF2122/97 INYRDEDFVARLKQETDGSGADIILDIMGASYLDRNIDALAT-DGQLIVI
Rv3777|M.tuberculosis_H37Rv INYRDEDFVARLKQETDGSGADIILDIMGASYLDRNIDALAT-DGQLIVI
MMAR_5332|M.marinum_M INYRDEDFVARLKQETDGSGADVILDIMGAAYLDRNVDALAV-DGQLVII
MUL_0096|M.ulcerans_Agy99 INYRDEDFVARLKQETDGSGADVILDIMGAAYLDRNVDALAV-DGQLVII
TH_1665|M.thermoresistible__bu ICYREEDFVERVRAETDGAGADVVLDIMGAAYLGRNVDALAT-DGRLVVI
MAB_0207c|M.abscessus_ATCC_199 INYRDTDFVQAVHSFTDGSGVDRILDIMGAKYLSRNVDALGN-DGHLVII
Mflv_2124|M.gilvum_PYR-GCK IDHTSTDYVDAVAEATRGRGVDVVFDTIGGDTLTRSPLSLAD-SGRVVSI
MSMEG_5762|M.smegmatis_MC2_155 VDLEHDRLEDVG-------EVDLVLDVIGGQILYRSAALVRP-TGSVVSI
Mvan_5758|M.vanbaalenii_PYR-1 IDYTATTVDEYVAAHTGGEGFDVVADNVGGATLDASFTAVKRYTGHVVSA
: * : * :*. * . : * ::
MLBr_00118|M.leprae_Br4923 GLQGGVKGELNLGKVLSKRARIIGSTLRARPVNGPNSKSEIVAAVTASIW
MAV_0242|M.avium_104 GMQGGVKAELNIGKLLAKRARVIGTTLRGRPVSGPNSKTEIVQAVTASVW
Mb3806|M.bovis_AF2122/97 GMQGGVKAELNLGKLLTKRARVIGTTLRARPVSGPHGKAAIAQAVAASVW
Rv3777|M.tuberculosis_H37Rv GMQGGVKAELNLGKLLTKRARVIGTTLRARPVSGPHGKAAIAQAVAASVW
MMAR_5332|M.marinum_M GMQGGIKAELNLGKLLTKRARVIGTTLRARPLQGPNGKAVIAQAVTASVW
MUL_0096|M.ulcerans_Agy99 GMQGGIKAELNLGKLLTKRARVIGTTLRARPLQGPNGKAVIAQAVTASVW
TH_1665|M.thermoresistible__bu GMQGGRRAELDLGKLVAKRAAVFGTALRGRPVTGPHGKADIVRAVVENVW
MAB_0207c|M.abscessus_ATCC_199 GMQGGVTAELNIAKLIAKRGSLTATTLRARPIAGPCGKAAIVESVAQNLW
Mflv_2124|M.gilvum_PYR-GCK VDI---ARPQNLIEAWGRNAAYHFVFTRQNRGK------------LDALT
MSMEG_5762|M.smegmatis_MC2_155 VEP---PRIP------ADNGRAVFFVVEPDRAG------------LVELE
Mvan_5758|M.vanbaalenii_PYR-1 LGWGTHSLAPLSFRGAAYSGVFTLMPLLTGRGRDHHGE------IVAEAA
.
MLBr_00118|M.leprae_Br4923 PMIADGSVRPIIG-ARMAVQQAADAHQLLLSGK---VSGKIVLTVAGSGT
MAV_0242|M.avium_104 PMIADGRVRPIIG-ARLPIQQAGDAHRRLVASE---VTGKIVLTV-----
Mb3806|M.bovis_AF2122/97 PMIAANRVRPVIG-TRLPIQQAAQAHELMLSGK---TFGKILLTV-----
Rv3777|M.tuberculosis_H37Rv PMIAANRVRPVIG-TRLPIQQAAQAHELMLSGK---TFGKILLTV-----
MMAR_5332|M.marinum_M PMIASNRVRPIIG-SRLPIGQAAQAHQLLLSGQ---SYGKTLLTVDG---
MUL_0096|M.ulcerans_Agy99 PMIASNRVRPIIG-SRLPIGQAAQAHQLLLSGQ---SYGKILLTVDG---
TH_1665|M.thermoresistible__bu PMVASDQVRPVVG-AEFPIQEAQAAHELLASGD---VYGKVLLCVED---
MAB_0207c|M.abscessus_ATCC_199 PLVEAGKVRPVVG-AEIPLSQAAEAHRLLENGA---VVGKVLLTR-----
Mflv_2124|M.gilvum_PYR-GCK MLVERGLVKPVLG-ATLPLARMGEAHELLENRRSYALRGKVAIDVAGETV
MSMEG_5762|M.smegmatis_MC2_155 QRLRDGRLRPRVG-TVYPLADAKEAFAPAQRPR-----GKSVIRVIEES-
Mvan_5758|M.vanbaalenii_PYR-1 ALADSGRLAPRLHPTTFPLDAVNDAHTAVESGS---ATGKVVVEPNV---
. : * : : .: *. ** :
MLBr_00118|M.leprae_Br4923 ETLLSRHH
MAV_0242|M.avium_104 --------
Mb3806|M.bovis_AF2122/97 --------
Rv3777|M.tuberculosis_H37Rv --------
MMAR_5332|M.marinum_M --------
MUL_0096|M.ulcerans_Agy99 --------
TH_1665|M.thermoresistible__bu --------
MAB_0207c|M.abscessus_ATCC_199 --------
Mflv_2124|M.gilvum_PYR-GCK ALPPRIS-
MSMEG_5762|M.smegmatis_MC2_155 --------
Mvan_5758|M.vanbaalenii_PYR-1 --------