For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KAIVAEQGAKGAGLSLTAVPYPHAAENDVIVQVHAAGFTPGELDWPGTWADRSGRSRGPSIPGHEVSGIV VELGYGTTGLTVGQRVFGITDWTRNGALAQYVAVEARNLAPLSADVDHTVAAALPVSGLTAWQGLFLHGH IVAGQSVLIHGAAGAVGSVAVQLAHEAGAHVIGTGRGGQRDQVLGLKADDFVDLTTDTLEDIGQVDVVFD VIGGEILDRSAQLVRAGGALITIAEPPRVIPEGARAVFFVVEADRRQLTILENKLRDGRIRQNIGSVIPL EDALSAFDIDRKTPGKTIVQIAAV*
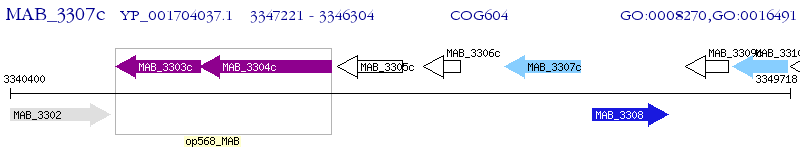
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3307c | - | - | 100% (305) | putative zinc-binding oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0750 | - | 2e-25 | 30.52% (308) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0207c | - | 1e-19 | 27.10% (321) | quinone oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3806 | - | 5e-24 | 28.45% (341) | putative oxidoreductase |
| M. gilvum PYR-GCK | Mflv_2124 | - | 1e-30 | 30.85% (295) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv3777 | - | 1e-23 | 28.15% (341) | oxidoreductase |
| M. leprae Br4923 | MLBr_00118 | - | 3e-22 | 26.19% (336) | putative oxidireductase |
| M. marinum M | MMAR_1738 | - | 7e-56 | 40.45% (309) | oxidoreductase |
| M. avium 104 | MAV_0242 | - | 1e-26 | 31.13% (318) | quinone oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_5762 | - | 1e-116 | 66.34% (303) | zinc-binding oxidoreductase |
| M. thermoresistible (build 8) | TH_1665 | - | 2e-24 | 30.53% (321) | PROBABLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_2935 | fadB5 | 3e-23 | 36.78% (174) | oxidoreductase FadB5 |
| M. vanbaalenii PYR-1 | Mvan_4202 | - | 3e-23 | 28.88% (329) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_2935|M.ulcerans_Agy99 ----MRAVVITKHGDPSVLQVQQRPDPPPPGPGQLRIAVRAAGVNF--AD
Mvan_4202|M.vanbaalenii_PYR-1 ----MRAAVCPQYGPPEVVRVEERPEPS-AGPGQVVVRVEAAAVNF--PD
Mb3806|M.bovis_AF2122/97 -MTIMRAVVAESSDR---LVWQEVPDVS-AGPGEVLIKVAASGVNR--AD
Rv3777|M.tuberculosis_H37Rv -MTIMRAVVAESSDR---LVWQEVPDVS-AGPGEVLIKVAASGVNR--AD
MLBr_00118|M.leprae_Br4923 ----MHAIVAESANQ---MVWREVPDVT-AGPGEVLIEVAASGVNR--AD
MAV_0242|M.avium_104 -MATMRAIVAESADQ---LSWQEVADVS-AGPGEVVVKVAAAGVNR--AD
TH_1665|M.thermoresistible__bu ----MRAISVQTSGQ---LSWVEVPDIE-PGESEVLIAVHTAGINR--AD
MAB_3307c|M.abscessus_ATCC_199 ----MKAIVAEQGAKG-AGLSLTAVPYPHAAENDVIVQVHAAGFTPGELD
MSMEG_5762|M.smegmatis_MC2_155 ----MRAIIAKDRTAGIDGLALAEVPYPHAAENDVIVAVHAAAFTPGELD
MMAR_1738|M.marinum_M ----MRALRAHR-RGGPETLAVERAPVPVPAAGEVVVAVRAAGITFDELT
Mflv_2124|M.gilvum_PYR-GCK MSDRMKAVVLAR-FGGADAFELRDVAVPHVGPRQVRVRVHATAVNP--LD
*:* . :: : * ::...
MUL_2935|M.ulcerans_Agy99 HLARVGLYPDAPKLPAVVGYEVAGTVEAAGAGVDESRIGERVLAGT----
Mvan_4202|M.vanbaalenii_PYR-1 VLLVADSYQVSVAPPFVPGSEFTGVVDEIGPHTTGFTVGERVAGTG----
Mb3806|M.bovis_AF2122/97 VLQAAGKYPPPPGVSDIIGLEVSGIVAAVGPGVTEWSAGQEVCALL----
Rv3777|M.tuberculosis_H37Rv VLQAAGKYPPPPGVSDIIGLEVSGIVAAVGPGVTEWSAGQEVCALL----
MLBr_00118|M.leprae_Br4923 VLQVAGKYPHPPGASAIIGMEVAGVVAAVSSGVTEWSVGQEVCALL----
MAV_0242|M.avium_104 VLQAAGKYPPPPGASETIGMEVSGVIAEVGDGVTEWSVGQEVCALL----
TH_1665|M.thermoresistible__bu LLQAAGKYPPPPGASEILGLEVSGTVAAVGAGVSGWTVGQPVCALL----
MAB_3307c|M.abscessus_ATCC_199 WPGTWADRSGRSRGPSIPGHEVSGIVVELGYGTTGLTVGQRVFGITDWTR
MSMEG_5762|M.smegmatis_MC2_155 WPGTWTDRAGRDRTPSVPGHEVSGVVAELGFGTTGLAVGQRVFGLTDWCR
MMAR_1738|M.marinum_M WDQTWT-RDGVSRTPVIPSHEVSGVIHEIGSGTEDFHCGDEVYGLIGFDR
Mflv_2124|M.gilvum_PYR-GCK YQIRRGDYTDLVPLPAIIGHDISGVIEEVGSHVTEFRAGDAVYYTPKIFG
. . :.:* : . . *: *
MUL_2935|M.ulcerans_Agy99 RFGGYSEIVNVTATDSVVLPEGLSFEQGAAVPVNYATAWAALHGYGSLRA
Mvan_4202|M.vanbaalenii_PYR-1 MFGAFAERVVADAAGLTPVPDGVDARTAAAFGVAHRTAYHALRSTARLSA
Mb3806|M.bovis_AF2122/97 AGGGYAEYVAVPADQVLPIPPSVNLVDSAALPEVACTVWSNLVMTAHLRP
Rv3777|M.tuberculosis_H37Rv AGGGYAEYVAVPADQVLPIPPSVNLVDSAALPEVACTVWSNLVMTAHLRP
MLBr_00118|M.leprae_Br4923 AGGGYAEYVAVPASQVMPIPKGVNVVDSAGIPEVACTVWSNLVMAAHLSA
MAV_0242|M.avium_104 AGGGYAEYVAVPAGQLLPIPAGVDLVDAAGLPEVACTVWSNLVLIAGLRN
TH_1665|M.thermoresistible__bu AGGGYAEYVAVPAEQVMPIPESVEIRYAAALPEVACTVWSNLVMTAGLSA
MAB_3307c|M.abscessus_ATCC_199 N-GALAQYVAVEARNLAPLSADVDHTVAAALPVSGLTAWQGLFLHGHIVA
MSMEG_5762|M.smegmatis_MC2_155 N-GTLAEYVAVEARNLAPLSATIDHTTAAALPISGLTAWQGLFTHGHLTP
MMAR_1738|M.marinum_M D-GAAADFVAVPAAHLAAKPATISHTVAAALPLAGLTALQALVDHAAVRP
Mflv_2124|M.gilvum_PYR-GCK GPGSYAEQHVADVDLVGRKPDNLDHLEAASLTLVGGTVWESLVTRAQLTV
* :: . . . :. .*.. *. * . :
MUL_2935|M.ulcerans_Agy99 GERVLVHAAAGGVGIAAVQFAKAAGAQVHGTASPGKHQRLA-ELGVDRAI
Mvan_4202|M.vanbaalenii_PYR-1 GDELVVLGAGGGVGLATVQLGVALGARVTAVASSNEKLAVAGGYGAATLI
Mb3806|M.bovis_AF2122/97 GQLVLIHGGASGIGSHAIQVARALAARVAITAGSPEKLELCRDLGAQITI
Rv3777|M.tuberculosis_H37Rv GQLVLIHGGASGIGSHAIQVVRALAARVAITAGSPEKLELCRDLGAQITI
MLBr_00118|M.leprae_Br4923 GQLLLLHGGTSGIGSHGIQVARALGAKVAVTAGSEEKLEFCRALGAQITI
MAV_0242|M.avium_104 GQLLLVHGGASGIGSHAIQVARALGARVAVTAGSAEKLGFCRELGADITI
TH_1665|M.thermoresistible__bu GEWVLIHGGASGVGSHAVQVARALDARVAVTAGTRHKLELCRELGAEVTI
MAB_3307c|M.abscessus_ATCC_199 GQSVLIHGAAGAVGSVAVQLAHEAGAHVIGTGRGGQ-RDQVLGLKADDFV
MSMEG_5762|M.smegmatis_MC2_155 GETVLIHGAAGGVGSIAVQLAREAGAHVIGSGRTAD-REYVLEVGAEAFV
MMAR_1738|M.marinum_M GEAVLVHGGAGGVGALVVQLAAILGANVTATVRSDA-GELVSRLGAQRVI
Mflv_2124|M.gilvum_PYR-GCK GETILIHGGAGGVGTIAIQVAKAMGARVITTAKAGD-HAFVNSLGADVAI
*: ::: .. ..:* :*. *.* . :
MUL_2935|M.ulcerans_Agy99 DYRRDGWWKGL------DSYDLVLDALGDTYLRRSYELLRPGG---RLVG
Mvan_4202|M.vanbaalenii_PYR-1 NHRDGELRQALRQAIP-DGADVVVDPVGGDVSEPALRSLGRGG---RFVS
Mb3806|M.bovis_AF2122/97 NYRDEDFVARLKQETDGSGADIILDIMGASYLDRNIDALATDG---QLIV
Rv3777|M.tuberculosis_H37Rv NYRDEDFVARLKQETDGSGADIILDIMGASYLDRNIDALATDG---QLIV
MLBr_00118|M.leprae_Br4923 NYRDEDFVARLQQQN--GGADVILDIMGAAYLDRNIDALAIDG---QLIV
MAV_0242|M.avium_104 NYRDEDFVARLADET--GGADVILDIMGAAYLDRNIDALATDG---QLVI
TH_1665|M.thermoresistible__bu CYREEDFVERVRAETDGAGADVVLDIMGAAYLGRNVDALATDG---RLVV
MAB_3307c|M.abscessus_ATCC_199 DLTTDTLEDIG-------QVDVVFDVIGGEILDRSAQLVRAGG---ALIT
MSMEG_5762|M.smegmatis_MC2_155 DLEHDRLEDVG-------EVDLVLDVIGGQILYRSAALVRPTG---SVVS
MMAR_1738|M.marinum_M DTRTEAFERTAT------DFDVVIDTVGGDTLDRSFAVLRPGG---RLVT
Mflv_2124|M.gilvum_PYR-GCK DHTSTDYVDAVAEATRGRGVDVVFDTIGGDTLTRSPLSLADSGRVVSIVD
*::.* :* : * .:
MUL_2935|M.ulcerans_Agy99 YGVSNMQEGEKRSLRRVAPHALSMLRGFNLMKQLEESKTVIGLNMLRLWD
Mvan_4202|M.vanbaalenii_PYR-1 VGFASG-----------------VIPRIPLNLVLIKGVHVLGFQFQDVPA
Mb3806|M.bovis_AF2122/97 IGMQGG-----------------VKAELNLGKLLTKRARVIGTTLRARPV
Rv3777|M.tuberculosis_H37Rv IGMQGG-----------------VKAELNLGKLLTKRARVIGTTLRARPV
MLBr_00118|M.leprae_Br4923 IGLQGG-----------------VKGELNLGKVLSKRARIIGSTLRARPV
MAV_0242|M.avium_104 IGMQGG-----------------VKAELNIGKLLAKRARVIGTTLRGRPV
TH_1665|M.thermoresistible__bu IGMQGG-----------------RRAELDLGKLVAKRAAVFGTALRGRPV
MAB_3307c|M.abscessus_ATCC_199 IAEPP--------------------------RVIPEGARAVFFVVEA---
MSMEG_5762|M.smegmatis_MC2_155 IVEPP--------------------------RIPADNGRAVFFVVEP---
MMAR_1738|M.marinum_M LSAPPP-----------------------AGKADEHGVTAMFFIVTP---
Mflv_2124|M.gilvum_PYR-GCK IARPQN-----------------------LIEAWGRNAAYHFVFTRQ---
MUL_2935|M.ulcerans_Agy99 D-----RGTLEPWIAPLSEALGQGTAAPIVHAAIPFAEAPEAHRILAA--
Mvan_4202|M.vanbaalenii_PYR-1 D-----EFTRN--EVELRELLVSGRVKPHVGAVYSLDDTVAALRHVSD--
Mb3806|M.bovis_AF2122/97 SGPHGKAAIAQAVAASVWPMIAANRVRPVIGTRLPIQQAAQAHELMLS--
Rv3777|M.tuberculosis_H37Rv SGPHGKAAIAQAVAASVWPMIAANRVRPVIGTRLPIQQAAQAHELMLS--
MLBr_00118|M.leprae_Br4923 NGPNSKSEIVAAVTASIWPMIADGSVRPIIGARMAVQQAADAHQLLLS--
MAV_0242|M.avium_104 SGPNSKTEIVQAVTASVWPMIADGRVRPIIGARLPIQQAGDAHRRLVA--
TH_1665|M.thermoresistible__bu TGPHGKADIVRAVVENVWPMVASDQVRPVVGAEFPIQEAQAAHELLAS--
MAB_3307c|M.abscessus_ATCC_199 ---------DRRQLTILENKLRDGRIRQNIGSVIPLEDALSAFDIDR---
MSMEG_5762|M.smegmatis_MC2_155 ---------DRAGLVELEQRLRDGRLRPRVGTVYPLADAKEAFAPAQ---
MMAR_1738|M.marinum_M ---------NRDQLNRLGELVGSGRLHVEIAQSFPLANGREAFESGRG--
Mflv_2124|M.gilvum_PYR-GCK ---------NRGKLDALTMLVERGLVKPVLGATLPLARMGEAHELLENRR
: : . : .. *
MUL_2935|M.ulcerans_Agy99 -RENIGKVVLVP-------------
Mvan_4202|M.vanbaalenii_PYR-1 -GRAIGKVVIDLS------------
Mb3806|M.bovis_AF2122/97 -GKTFGKILLTV-------------
Rv3777|M.tuberculosis_H37Rv -GKTFGKILLTV-------------
MLBr_00118|M.leprae_Br4923 -GKVSGKIVLTVAGSGTETLLSRHH
MAV_0242|M.avium_104 -SEVTGKIVLTV-------------
TH_1665|M.thermoresistible__bu -GDVYGKVLLCVED-----------
MAB_3307c|M.abscessus_ATCC_199 --KTPGKTIVQIAAV----------
MSMEG_5762|M.smegmatis_MC2_155 --RPRGKSVIRVIEES---------
MMAR_1738|M.marinum_M AARRAGKTVITVGQ-----------
Mflv_2124|M.gilvum_PYR-GCK SYALRGKVAIDVAGETVALPPRIS-
** :