For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTSTATEPITYGSQRLAELIEKIGEGSLERERTGERPFAVIDLIKQSGLGALRVPREEGGGGATLRDLFA TVIAIATADVNVAHILRGHFAHIEERLRLDSELRQKGIDLALSGKLIGNASTEIGSTPAGGFTWQTRLTR DGDNYLLNGKKFFTTGTLYSDYAEVLASDPTGASVITLVPTDHPGVTVLDDWDGMGQRATGTGTAIFENV PVAAEEVMEFAPPGADDEEQSLYHSGAFFQLYVTALEVGVIHALRRDAVEHVHTRARSFAWAPHPVPADD PLLQREIGEIAAAAYAGEATVLAAAEALGRAYQADIDKTDPDLVLAHEASLQAAQAKIVVDALAQKAASQ IFDVGGASIVRQAYLLDRHWRNIRTLASHNPSSYKAQAVGAFFTRGTRLPGSGYF
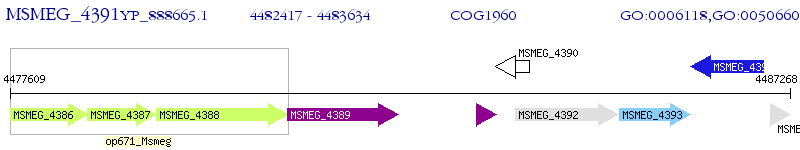
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4391 | - | - | 100% (405) | acyl-CoA dehydrogenase-family protein |
| M. smegmatis MC2 155 | MSMEG_5001 | - | 5e-80 | 41.21% (398) | pesticide degrading monooxygenase |
| M. smegmatis MC2 155 | MSMEG_4084 | - | 5e-76 | 40.15% (391) | putative acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2812c | fadE21 | 9e-11 | 23.77% (265) | acyl-CoA dehydrogenase FADE21 |
| M. gilvum PYR-GCK | Mflv_4548 | - | 8e-12 | 23.24% (383) | acyl-CoA dehydrogenase type 2 |
| M. tuberculosis H37Rv | Rv2789c | fadE21 | 9e-11 | 23.77% (265) | acyl-CoA dehydrogenase FADE21 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 2e-06 | 24.27% (206) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1422c | - | 2e-87 | 43.80% (395) | putative acyl-CoA dehydrogenase |
| M. marinum M | MMAR_4030 | - | 3e-09 | 22.07% (367) | acyl-CoA dehydrogenase |
| M. avium 104 | MAV_3422 | - | 5e-72 | 41.49% (376) | acyl-CoA dehydrogenase domain-containing protein |
| M. thermoresistible (build 8) | TH_3393 | - | 3e-50 | 35.68% (426) | PUTATIVE Acyl-CoA dehydrogenase, type 2, C-terminal domain |
| M. ulcerans Agy99 | MUL_2144 | fadE21 | 3e-09 | 24.89% (225) | acyl-CoA dehydrogenase FadE21 |
| M. vanbaalenii PYR-1 | Mvan_3173 | - | 7e-39 | 31.84% (402) | acyl-CoA dehydrogenase type 2 |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_1422c|M.abscessus_ATCC_199 --------MTSAPSVLEIRALPGTDTYRNLLAQVAEGARDRDLD--DENP
MAV_3422|M.avium_104 --------MPVAFHRQDT--PAGAAAVQPVLAEIAADYRLRLADGGTEPP
MSMEG_4391|M.smegmatis_MC2_155 --------MTSTATEPIT---YGSQRLAELIEKIGEGSLERERTG--ERP
TH_3393|M.thermoresistible__bu -------VTEPLTMARIVDTAEALAVAARLSGSFAAEASTRDAER--LLP
Mvan_3173|M.vanbaalenii_PYR-1 ---MATDITDRIPDTLLRTPADALAAAEEVAEAIAAGSAERERAG--AVL
Mb2812c|M.bovis_AF2122/97 -----------MFEWSDTDLMVRDAVRQFIDKEIRPHQDALETGE--LSP
Rv2789c|M.tuberculosis_H37Rv -----------MFEWSDTDLMVRDAVRQFIDKEIRPHQDALETGE--LSP
MUL_2144|M.ulcerans_Agy99 -----------MIEWSDTDLMMRGAVRQFIDREIRPNLDELETGA--LSP
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRA--RFP
MMAR_4030|M.marinum_M -----MTKLAQTLGLTELQAEIVATVRQFVDKEVIPNASELERKD--AYP
Mflv_4548|M.gilvum_PYR-GCK ------------------MSERVLDRLNELAGQFKEQAVEAEKLG--KLP
.
MAB_1422c|M.abscessus_ATCC_199 FDQVRLLKEAGFGRLRIPTELG---------------------GAGLSVR
MAV_3422|M.avium_104 LRGLKLVRDHRLGALRLARELG---------------------GAGYTAP
MSMEG_4391|M.smegmatis_MC2_155 FAVIDLIKQSGLGALRVPREEG---------------------GGGATLR
TH_3393|M.thermoresistible__bu YEQVCELKRSGLLTLSVPAEHG---------------------GLDVPAT
Mvan_3173|M.vanbaalenii_PYR-1 TPQLRLVAESGLLGISVPTAHG---------------------GPGLPAS
Mb2812c|M.bovis_AF2122/97 YPIARKLFSQFGLDVLLAESVNQMLDGERAKR-EKRD-SSGSFGLADQAS
Rv2789c|M.tuberculosis_H37Rv YPIARKLFSQFGLDVLLAESVNQMLDGERAKR-EKRD-SSGSFGLADQAS
MUL_2144|M.ulcerans_Agy99 YPIMRKLFSQFGLDVLAAESVKSILDAERANQDEKRHRTSGSGGLGPQAS
MLBr_00737|M.leprae_Br4923 EEALAALNASGFNAIHVPEEYG---------------------GQGADSV
MMAR_4030|M.marinum_M HEIVEQMRSIGLFGLMIPEQYG---------------------GLGESLL
Mflv_4548|M.gilvum_PYR-GCK DATVKSMKSIGSIRLLQPKKHG---------------------GLEVHPR
: : . *
MAB_1422c|M.abscessus_ATCC_199 QLFSAIIDVAQADPIVAHIFRTHFWFVEERLRTLADPV-SAAWLTKVNEG
MAV_3422|M.avium_104 EFFDYLVALAAADPDLAHILRIHYALVEELQVTPRRPG-SDRWIAVVAEG
MSMEG_4391|M.smegmatis_MC2_155 DLFATVIAIATADVNVAHILRGHFAHIEERLR-LDSEL-RQKGIDLALSG
TH_3393|M.thermoresistible__bu VLAEVFRLLAHGDPSVAQIPHSHFTFLEVLRLQGTERQ-RGFFYGLVLEG
Mvan_3173|M.vanbaalenii_PYR-1 TVVEVLRRLARADGAVGQLLLSHFVVAQAISGLGHTQP-APRIYADLLAG
Mb2812c|M.bovis_AF2122/97 MVAVLVSELAGVSIGLLSTVAVSLGLGAATIMSRGTLAQQERWVPTLVTL
Rv2789c|M.tuberculosis_H37Rv MVAVLVSELAGVSIGLLSTVAVSLGLGAATIMSRGTLAQQERWVPTLVTL
MUL_2144|M.ulcerans_Agy99 MAAVLVSELAGVSIGLLSTVAVSLGLGAATIMSRGTLAQKQRWLPELMTL
MLBr_00737|M.leprae_Br4923 AACIVIEEVARVDASASLIPAVNK-LGTMGLILRGSEELKKQVLPSLAAE
MMAR_4030|M.marinum_M TYALCVEELARGWMSVSGVLNTHF-IVAYMLRQHGTEEQRQRFLPRMATG
Mflv_4548|M.gilvum_PYR-GCK EFAETVMATAALDPAAGWVN----------------GVVGVHPYQLAYAD
. *
MAB_1422c|M.abscessus_ATCC_199 NLFANAFS--EKGSLAVGSLVFNTRLLPVGDAYRLNGEKFYSTGTLFADY
MAV_3422|M.avium_104 GLIGGANA--EQSQKSVGGNHRDTRLVTTPDGLRLRGRKFYSTGAQFSDY
MSMEG_4391|M.smegmatis_MC2_155 KLIGNAST--EIGSTPAGGFTWQTRLTRDGDNYLLNGKKFFTTGTLYSDY
TH_3393|M.thermoresistible__bu ALFANAQS--ERG-PHPIDVDTTVLLRRGAGDYLLTGRKFYSTGALFADW
Mvan_3173|M.vanbaalenii_PYR-1 AQLGNASA--ERGTAHALDRRTTVTRRD--GHWVLNGTKYYATGTLGATW
Mb2812c|M.bovis_AF2122/97 EKIAAWAITEPDSGSDAFGGMKTHVTRDGE-DYILNGHKTFITNGPYADV
Rv2789c|M.tuberculosis_H37Rv EKIAAWAITEPDSGSDAFGGMKTHVTRDGE-DYILNGHKTFITNGPYADV
MUL_2144|M.ulcerans_Agy99 EKIAAWAITEPDSGSDAFGGMKTHVKRDGA-DYILNGQKTFITNGPYADV
MLBr_00737|M.leprae_Br4923 GAMASYALSEREAGSDAAS-MRTRAKADGD-DWILNGFKCWITNGGKSTW
MMAR_4030|M.marinum_M ESRGAFSMSEPELGSDVAA-IRTRATRGADGNYTIDGQKMWLTNGGSSTL
Mflv_4548|M.gilvum_PYR-GCK PKVGEEIWADDIDTWVASPYAPQGVAVPTDGGYIFTGRWQFSSGTDACDW
. : * : :. .
MAB_1422c|M.abscessus_ATCC_199 LTVTATTDRD---------SVASVIVPGDR-----DGVRL-VDDWDGFGQ
MAV_3422|M.avium_104 LRITAQDDDG---------APTAVVIPVDR-----AGVEH-LDDWDGIGQ
MSMEG_4391|M.smegmatis_MC2_155 AEVLASDPTG---------ASVITLVPTDH-----PGVTV-LDDWDGMGQ
TH_3393|M.thermoresistible__bu IVVRASVPADAADAPTAATPKAVAFVPRHA-----PGLTV-IDDWDGMGQ
Mvan_3173|M.vanbaalenii_PYR-1 IAVAARIEGRENGD------TATAFVRPDQ-----EGVTLDLESWSAFGQ
Mb2812c|M.bovis_AF2122/97 LVVYAKLAD-GEPASDWRNRPVLVFVLDAG----MPGLTQ-GKPFKKMGM
Rv2789c|M.tuberculosis_H37Rv LVVYAKLAD-GEPASDWRNRPVLVFVLDAG----MPGLTQ-GKPFKKMGM
MUL_2144|M.ulcerans_Agy99 LLVYAKLDEAGATSSDRRDRPVLIFVLESG----MAGLTQ-GKPFKKMGM
MLBr_00737|M.leprae_Br4923 YTVMAVTDP--DKGAN--GISAFIVHKDD------EGFSI-GPKEKKLGI
MMAR_4030|M.marinum_M VAVLVRTDEGAAQPHR--NLTAFLVEKPTGFGEVLPGLTI-PGKIDKLGY
Mflv_4548|M.gilvum_PYR-GCK IILGAMVGDAKGVPLMPPQMLHMILPRK--------DYQIVDDSWDVVGL
: . . . . .*
MAB_1422c|M.abscessus_ATCC_199 RRTGTGTTIFSQVDVAADEILSETPY-DAAPVPT---TQYASLQLYILAI
MAV_3422|M.avium_104 RQTGSGTTVFNDVAVADDEVFPLG---STIGVNR---ARGALVQLYLHAV
MSMEG_4391|M.smegmatis_MC2_155 RATGTGTAIFENVPVAAEEVMEFAPP-GADDEEQSLYHSGAFFQLYVTAL
TH_3393|M.thermoresistible__bu RTTASGSVTLDEVRVPADHVV------PFTPIFRRPTVYGARAQLWHAAL
Mvan_3173|M.vanbaalenii_PYR-1 RGTASGEVRLNNVVVDDDLLIDEGP--PPDPITADPSVLGAFDQALHAAI
Mb2812c|M.bovis_AF2122/97 MSSPTGELFFDNVRLTPDRLLCAE-----GDGRDSARANFAVERLGVALM
Rv2789c|M.tuberculosis_H37Rv MSSPTGELFFDNVRLTPDRLLCAE-----GDGRDSARANFAVERLGVALM
MUL_2144|M.ulcerans_Agy99 MSSPTGELFFDNVRLTPDRLLGESEHHGDGDGRESARANFAVERLGVALM
MLBr_00737|M.leprae_Br4923 KGSPTTELYFDKCRIPGDRIIG-EP----GTGFKTALATLDHTRPTIGAQ
MMAR_4030|M.marinum_M KGIDTTELIFDGYRATADDILGGTP----GQGFFQMMDGIEVGRVNVSAR
Mflv_4548|M.gilvum_PYR-GCK RGTGSKDVIVDGAFVPTYRTMDGMKVMDGSAQIEAGMTEPLYKMPWSTMF
: .. .
MAB_1422c|M.abscessus_ATCC_199 VAGVLRAVVDDGEALLRSRQRN---FSHALTERPVDDPLLQRTLGELSAT
MAV_3422|M.avium_104 AAGILRSLTEDAAALVRGRRRT---YTFASADTPTADPQLLQIVGEIDAV
MSMEG_4391|M.smegmatis_MC2_155 EVGVIHALRRDAVEHVHTRARS---FAWAPHPVPADDPLLQREIGEIAAA
TH_3393|M.thermoresistible__bu DVGIATAALEEG-VRLAARARP---HFESGAPVATDDPTLIQSAGEVAVT
Mvan_3173|M.vanbaalenii_PYR-1 DIGIARAALEDGAVFVRTRSRPWKEALDAGIERADQEPHVVRRFGELTAR
Mb2812c|M.bovis_AF2122/97 SLGIINECHRLCVDYAKTRTLWG--------RNIGQFQLIQLKLAKMEVA
Rv2789c|M.tuberculosis_H37Rv SLGIINECHRLCVDYAKTRTLWG--------RNIGQFQLIQLKLAKMEVA
MUL_2144|M.ulcerans_Agy99 ALGIINECHRLCVDYAKTRTLWG--------KNIGQFQLIQLKLAKMEVA
MLBr_00737|M.leprae_Br4923 AVGIAQGALDAAIVYTKDRKQFG--------ESISTFQSIQFMLADMAMK
MMAR_4030|M.marinum_M ACGVGLRAFELAVRYAQQRHTFG--------KPIAEHQAISFQLAEMATK
Mflv_4548|M.gilvum_PYR-GCK PLGISSATIGICEGALAAHLDYQRERVGANGTAIKDDPYVMYAIGDAAAE
*: : : ..
MAB_1422c|M.abscessus_ATCC_199 AFAAEATVLAAADAIEAATATLRD-GVPDAQLAQEAQVAVAKAKVHIDAV
MAV_3422|M.avium_104 AYTAHALVRNAAAELAPALQAARD-TGIDPELEAAASIAAARVKVAIQEP
MSMEG_4391|M.smegmatis_MC2_155 AYAGEATVLAAAEALGRAYQADIDKTDPDLVLAHEASLQAAQAKIVVDAL
TH_3393|M.thermoresistible__bu VRAAQALLGEAARAVD----AATAD--LTEDTAAAASIAVATAKVAAERA
Mvan_3173|M.vanbaalenii_PYR-1 LYALEALLARGTALID----EAYAAPEVTRDAAAQASLTVAAAKALAQQD
Mb2812c|M.bovis_AF2122/97 RINVQNMVFQAIERLK--------------AGKQLTLAEASAIKLYSSEA
Rv2789c|M.tuberculosis_H37Rv RINVQNMVFQAIERLK--------------AGKQLTLAEASAIKLYSSEA
MUL_2144|M.ulcerans_Agy99 RINVENMVFQTLEKLK--------------AGREPTLAEASAIKLYSSEA
MLBr_00737|M.leprae_Br4923 VEAARLIVYAAAARAE--------------RGEPDLGFISAASKCFASDI
MMAR_4030|M.marinum_M VEAAHLMMVHAARLKD--------------SGERND-VAAGMAKYLASEY
Mflv_4548|M.gilvum_PYR-GCK IEASRNTLLSNVDRVY-------DIVDSGREVTFETRAAVRRSQIKAVWR
. : :
MAB_1422c|M.abscessus_ATCC_199 ALDAATKLLDLGGASAASRTRNLDRHWRNIRT---ISLHNPVH-YKARVI
MAV_3422|M.avium_104 ALRAASRVFDAGGASAVQASALLDRHWRNLRT---LFSHNPTV-YKARVL
MSMEG_4391|M.smegmatis_MC2_155 AQKAASQIFDVGGASIVRQAYLLDRHWRNIRT---LASHNPSS-YKAQAV
TH_3393|M.thermoresistible__bu ALRASTALFELGGTRSATVSANLSRFWRDART---HTLHDPTR-WKLHHI
Mvan_3173|M.vanbaalenii_PYR-1 AVEIAGAIFELTGTSGTDGAVNLDRHWRNVRV---HSLHDPAR-WKYVHL
Mb2812c|M.bovis_AF2122/97 ATDVAMEAVQLFGGNGYMAEYRVEQLARDAKS---LMIYAGSNEVQVTHI
Rv2789c|M.tuberculosis_H37Rv ATDVAMEAVQLFGGNGYMAEYRVEQLARDAKS---LMIYAGSNEVQVTHI
MUL_2144|M.ulcerans_Agy99 ATDVAMEAVQLFGGNGYMAEYRVEQLARDAKS---LMIYAGSNEVQVTHI
MLBr_00737|M.leprae_Br4923 AMEVTTDAVQLFGGAGYTSDFPVERFMRDAKI---TQIYEGTNQIQRVVM
MMAR_4030|M.marinum_M CSEVTQQSFRIHGGYGYSKEYEIERLMRDAPF---LLIGEGTSEIQKSII
Mflv_4548|M.gilvum_PYR-GCK AVAAVDQIFARSGGNALRMDKPLQRYWRDAHAGVAHAIHVPGTVYHASAL
. . * :.: *: : :
MAB_1422c|M.abscessus_ATCC_199 GQNLLHGTPLPANAYF-
MAV_3422|M.avium_104 GDVVVNGAALPDTSFF-
MSMEG_4391|M.smegmatis_MC2_155 GAFFTRGTRLPGSGYF-
TH_3393|M.thermoresistible__bu GRYLLSGTRPPRHGQL-
Mvan_3173|M.vanbaalenii_PYR-1 GNHTLHDTRPPRLGLIL
Mb2812c|M.bovis_AF2122/97 AKGLLGEPASRA-----
Rv2789c|M.tuberculosis_H37Rv AKGLLGEPASRA-----
MUL_2144|M.ulcerans_Agy99 AKGLLG-----------
MLBr_00737|M.leprae_Br4923 SRALLR-----------
MMAR_4030|M.marinum_M SKRLLHEYRE-------
Mflv_4548|M.gilvum_PYR-GCK SSLGVDPQGPLRAMI--
.