For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSHSQIFPTPTDTRTISAALVRATGEPFVITDLALTGPRADEILVRVDAVGICHADTQAQTGALPVPAPM VAGHEGTGIVEEVGSDVQHLKCGDRVILTFDSCGGCDRCRSGIPTQCREFVNRNFTAGARPDGTARLHLD TNAVNGSFFGQSSFATYALATERNAVAVSTQLPPELLAPVGCAIQTGVGTVLDVLAPPPAAPVVVFGAGA VGLSAVMAAAATGHEVVAVDTKPERLDLARKLGATHVVDPARADVTRAITAIMPGGAPHVFESSGAPVAL IAGVEALAPGGTIAVVGTSAAGVTVGIDVVDLVNKSKHIVGVVEGASRPHESIPRLVKMIEDGALPVHEI VTTFPLEDIAAAIDAAKSGRVVKPVLLPH
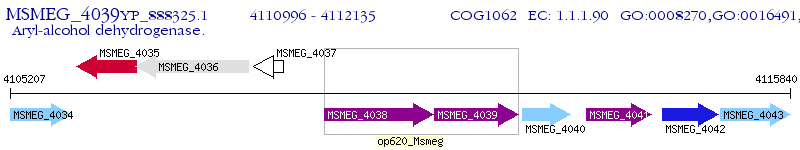
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4039 | - | - | 100% (379) | aryl-alcohol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2034 | - | 6e-75 | 46.41% (362) | aryl-alcohol dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1138 | - | 4e-59 | 36.17% (376) | alcohol dehydrogenase 1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0167c | adhE1 | 4e-52 | 35.59% (354) | putative zinc-type alcohol dehydrogenase (e subunit) adhE |
| M. gilvum PYR-GCK | Mflv_0725 | - | 8e-75 | 43.49% (361) | alcohol dehydrogenase |
| M. tuberculosis H37Rv | Rv0162c | adhE1 | 4e-52 | 35.59% (354) | zinc-type alcohol dehydrogenase E subunit |
| M. leprae Br4923 | MLBr_01784 | adhE2 | 9e-36 | 26.83% (369) | putative alcohol dehydrogenase (Zn dependent) |
| M. abscessus ATCC 19977 | MAB_4388 | - | 3e-72 | 42.70% (363) | zinc-containing alcohol dehydrogenase |
| M. marinum M | MMAR_0405 | adhE | 9e-51 | 34.92% (358) | zinc-type alcohol dehydrogenase (E subunit) AdhE |
| M. avium 104 | MAV_5022 | - | 9e-54 | 37.36% (364) | oxidoreductase, zinc-binding dehydrogenase family protein |
| M. thermoresistible (build 8) | TH_2558 | - | 1e-75 | 41.55% (361) | PUTATIVE putative aryl-alcohol dehydrogenase |
| M. ulcerans Agy99 | MUL_3861 | adhB_1 | 9e-39 | 29.66% (381) | zinc-containing alcohol dehydrogenase NAD dependent AdhB_1 |
| M. vanbaalenii PYR-1 | Mvan_5270 | - | 4e-40 | 29.29% (379) | alcohol dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0167c|M.bovis_AF2122/97 -MPAVQPWLYSNMPAIRGAVLDQIGVPRPYWRSKPISVVELHLDPPDRGE
Rv0162c|M.tuberculosis_H37Rv -MPAVQPWLYSNMPAIRGAVLDQIGVPRPYWRSKPISVVELHLDPPDRGE
MMAR_0405|M.marinum_M MELGLQPWLYSNMIHIRGAVLERIGSPRPYAQSRPISVVDLDLDPPGDDE
MAV_5022|M.avium_104 -------------------MLQRIGAPRPYARSRPISITDVELAPPGRDE
Mflv_0725|M.gilvum_PYR-GCK --------------------MPETTAVLSYAPEDPFSVAEVRVDDPRADE
MAB_4388|M.abscessus_ATCC_1997 ---------------------MKVTAALSASPAAPFTLQELELDEPRSDE
TH_2558|M.thermoresistible__bu ---------------------------VLRGETDPYQIEELDLPDPAAHQ
MSMEG_4039|M.smegmatis_MC2_155 --------MSHSQIFPTPTDTRTISAALVRATGEPFVITDLALTGPRADE
MUL_3861|M.ulcerans_Agy99 ---------------------MRSRAAILHDVGEPWSVEEFELDAPRTAE
Mvan_5270|M.vanbaalenii_PYR-1 ---------------------MKTNAAILWEYGGDWTVEEIDLDPPGDGE
MLBr_01784|M.leprae_Br4923 -------------------MSQTVRGVISRKKDEPVELVDIVIPDPGPGE
: :. : * :
Mb0167c|M.bovis_AF2122/97 VLVRIEAAGVCHSDLSVVDGTRVRP------------VPILLGHEAAGIV
Rv0162c|M.tuberculosis_H37Rv VLVRIEAAGVCHSDLSVVDGTRVRP------------VPILLGHEAAGIV
MMAR_0405|M.marinum_M VLVRIEAAGVCHSDLSVVDGNRVRP------------VPMLLGHEAAGIV
MAV_5022|M.avium_104 VLVRIEAAGICHSDLSVVDGNRVRP------------VPMLLGHEAAGIV
Mflv_0725|M.gilvum_PYR-GCK ILVRIEAAGVCHTDLVNRVAGKP-D------------RPVLLGHEGAGIV
MAB_4388|M.abscessus_ATCC_1997 VLVKIHATGLCHTDLT--FKSQV-P------------IPAVLGHEGAGIV
TH_2558|M.thermoresistible__bu ILVRIAGTGHCHTDVLPRAGAGFGT------------PPIVVGHEGSGVV
MSMEG_4039|M.smegmatis_MC2_155 ILVRVDAVGICHADTQAQTGALPVP------------APMVAGHEGTGIV
MUL_3861|M.ulcerans_Agy99 VLVEMSAAGLCHTDDHILKGDMSAPNEVLRSRGLPTMFPTIGGHEGSGIV
Mvan_5270|M.vanbaalenii_PYR-1 VLVSWEATGLCHSDEHIRTGDLPAP------------LPLIGGHEGAGIV
MLBr_01784|M.leprae_Br4923 AVVDVTACGVCHTDLTYREGGINDR------------YPFLLGHEAAGTV
:* . * **:* * : ***.:* *
Mb0167c|M.bovis_AF2122/97 EQVGDGVDGVAVGQRVVLVFLPRCGQCAACATDGRTPCEPGSAAN-KAGT
Rv0162c|M.tuberculosis_H37Rv EQVGDGVDGVAVGQRVVLVFLPRCGQCAACATDGRTPCEPGSAAN-KAGT
MMAR_0405|M.marinum_M ERVGDRVDDLAVGQRVVLVFLPRCGHCPSCATEGITPCAPGSATN-AAGT
MAV_5022|M.avium_104 ERVGPGVADLAVGARVVLVFLPRCGRCAACATEGLTPCEPGSAAN-GAGT
Mflv_0725|M.gilvum_PYR-GCK ERVGDGLQSVKPGDRVVLSFRS-CGACTNCRARRPAYCRKAVRLN-QFGQ
MAB_4388|M.abscessus_ATCC_1997 TAVGEAVSGISPGDHVVLSYRS-CGGCRSCAAGERAYCSRSMRLN-SAGV
TH_2558|M.thermoresistible__bu EAVGAAVDTVAVGDHVVLSFDS-CGTCGSCLAAHPAYCDTFLERN-LGGP
MSMEG_4039|M.smegmatis_MC2_155 EEVGSDVQHLKCGDRVILTFDS-CGGCDRCRSGIPTQCREFVNRNFTAGA
MUL_3861|M.ulcerans_Agy99 REIGPGVTDFVPGDHVVMSFVAVCGQCRWCASGMEYLCDAGIGTLTPGMP
Mvan_5270|M.vanbaalenii_PYR-1 REVGPGVVGLAPGDHVVASFLPACGRCRFCSTGHQNLCDLG-AMIMAGTQ
MLBr_01784|M.leprae_Br4923 EVVGPGVTAVEPGDFVILNWRAVCGQCRACKRGRPSYCFDTFNAQQKMTL
:* : . * *: : . ** * * *
Mb0167c|M.bovis_AF2122/97 LLGGGIRLS-RGGRPVY-HHLGVSGFATHVVVNRASVVPVPHEVPPTVAA
Rv0162c|M.tuberculosis_H37Rv LLGGGIRLS-RGGRPVY-HHLGVSGFATHVVVNRASVVPVPHEVPPTVAA
MMAR_0405|M.marinum_M LLGGGIRIS-RSGQPIY-HHLGVSGFATHAVVNRASAVAVPDEVPPQVAA
MAV_5022|M.avium_104 LLGGDIRLS-RAGRPVY-HHLGVSGFATHAVVNRASAVEVPPDVPAEVAA
Mflv_0725|M.gilvum_PYR-GCK RADGTPRVT-VDGRSVLDGFFGQSSLAGYALTTQDNTVVVGPDVDPVLAA
MAB_4388|M.abscessus_ATCC_1997 RADGSPTLS-DNGTPVYGSFFGQSSFAQYALTGADNTVVVDPSIDLAVAA
TH_2558|M.thermoresistible__bu GVDGAAQLTDVDGQPVAGRWFGQSSFASHAVVDAANAVVVDKDLPLELLG
MSMEG_4039|M.smegmatis_MC2_155 RPDGTARLH-LDTNAVNGSFFGQSSFATYALATERNAVAVSTQLPPELLA
MUL_3861|M.ulcerans_Agy99 TDGTFRHHT--ADGTPLGHLAKVGAFAEHTVVSRDSLVKIDPDLPLAPSA
Mvan_5270|M.vanbaalenii_PYR-1 LDGTYRRHV--R-GQDVGVMALVGTFSQYGTVPEASIVKIDDDIPLSRAC
MLBr_01784|M.leprae_Br4923 IDG-----------TELTPALGIGAFADKTLVHSGQCTKVDPAADPAVAG
. . :: . . . :
Mb0167c|M.bovis_AF2122/97 LLGCAVLTGGGAVLNVGDPQPGQSVAVVGLGGVGMAAVLTALTYTDVRVV
Rv0162c|M.tuberculosis_H37Rv LLGCAVLTGGGAVLNVGDPQPGQSVAVVGLGGVGMAAVLTALTYTDVRVV
MMAR_0405|M.marinum_M LLGCAVLTGGGAVLNVGKPVAGQSVAVVGLGGVGMAAVLTALTQDGVAVF
MAV_5022|M.avium_104 LLGCAVLTGGGAVLNVGRPAPGQTVAVVGLGGVGMAALLTALTYDGVPVV
Mflv_0725|M.gilvum_PYR-GCK PLGCGFQTGAGAVLNLMQPAAGSSVAVFGAGAVGLAGVLAALAVDVETVI
MAB_4388|M.abscessus_ATCC_1997 PLGCGFQTGAGAVLNVLSPEPDSRLVVFGAGGVGLAAVMAAKAQGVETII
TH_2558|M.thermoresistible__bu PLGCGVQTGAGAVLEVLRIGPGESIVITGTGAVGLSAVMAAKVAGASTII
MSMEG_4039|M.smegmatis_MC2_155 PVGCAIQTGVGTVLDVLAPPPAAPVVVFGAGAVGLSAVMAAAATG-HEVV
MUL_3861|M.ulcerans_Agy99 LLSCAIPTGYGSAANRAGVRGGDTVVVLGAGGIGTGAIQGARINGAAQIV
Mvan_5270|M.vanbaalenii_PYR-1 LLGCGVTTGWGSAVNTADVSPGDTVVIIGLGGIGSGAIQGARLAGAEKIV
MLBr_01784|M.leprae_Br4923 LLGCGVMAGLGAAINTAAVSRDDTVAVIGCGGVGDAAISGAALVGANRII
:.*.. :* *:. : :.: * *.:* ..: * :.
Mb0167c|M.bovis_AF2122/97 AVDQLPEKLSAAKALGAHEIYTPQQATAGGVK--------AAVVVEAVGH
Rv0162c|M.tuberculosis_H37Rv AVDQLPEKLSAAKALGAHEIYTPQQATAGGVK--------AAVVVEAVGH
MMAR_0405|M.marinum_M GVDQLPQKLSAAEALGASATYTPQQAAAAGVK--------ADVVIEAAGH
MAV_5022|M.avium_104 AVDQLPEKLAAARRLGAHQAYTPQQAVEARVK--------AAVVIEAVGH
Mflv_0725|M.gilvum_PYR-GCK VVEPSPQRRALAESLGVRAALSPDD-HTVRAIREITG-GGVDYALDTTGR
MAB_4388|M.abscessus_ATCC_1997 AVDPVASRRAKATELGATQTVDPAM-EDVAAM----A-RGATHALDTTAN
TH_2558|M.thermoresistible__bu AVDLNTERLALAEEFGATHTIVANTGELTEQIRAIVP-GGTHYGLDTTGL
MSMEG_4039|M.smegmatis_MC2_155 AVDTKPERLDLARKLGATHVVDPARADVTRAITAIMP-GGAPHVFESSGA
MUL_3861|M.ulcerans_Agy99 AIDPVEFKQKSAYLFGATHGAGSIS-QAMALVRELTHGVMADSVLVSPSL
Mvan_5270|M.vanbaalenii_PYR-1 VIEPVESKRELAFRFGATHFVTSME-EATALVADLTRGVMADSAILTVGL
MLBr_01784|M.leprae_Br4923 AVDIDDTKLEWARTFGATHTVNALELEVVKTIQDLTSGFGVDVVIDTVGR
:: : * :*. . . . : .
Mb0167c|M.bovis_AF2122/97 PA--ALHTAIGLTAPGGRTITVGL-PPPDVRISLSPLDFVTEGRSLIGSY
Rv0162c|M.tuberculosis_H37Rv PA--ALHTAIGLTAPGGRTITVGL-PPPDVRISLSPLDFVTEGRSLIGSY
MMAR_0405|M.marinum_M PA--ALETAIGLTAPGGRTITVGL-PPPDARISVAPLGFVAEGRSLIGSY
MAV_5022|M.avium_104 PA--ALQTAIDLTAPGGRTITVGL-PPPDARITVSPLGFVAEGRSLIGSY
Mflv_0725|M.gilvum_PYR-GCK PE--VLASAVAALAVGGTAVAVGL-GTG--VPAIDLADLVLRGKTVHGCL
MAB_4388|M.abscessus_ATCC_1997 AG--VIATALGLLRQRGVLVLVGL-GAQ--PGPIDLNDLMLGGKVIRGCI
TH_2558|M.thermoresistible__bu PA--VIAAALEAVAVRGTMGLVGV-QQG--ELAIGPLALTIG-RTITGIL
MSMEG_4039|M.smegmatis_MC2_155 PV--ALIAGVEALAPGGTIAVVGT-SAAGVTVGIDVVDLVNKSKHIVGVV
MUL_3861|M.ulcerans_Agy99 ITADDVRDALRLTCKGGTCVLAGMTSQLTRSVNIDLQEFILMNKTLAGTI
Mvan_5270|M.vanbaalenii_PYR-1 LEGSMLEEAANIIRKGGAVVMTALSTMADNQPTLGMMMFTLFQKRLLGSL
MLBr_01784|M.leprae_Br4923 PE--TWKQAFYARDLAGTVVLVGVPTPDMR-LDMPLLDFFSHGGSLKSSW
. * .. : : : .
Mb0167c|M.bovis_AF2122/97 LGSAVPSHDIPRFVSLWQSGRLPVESLVTSTIRLDDINEAMDHLADGIAV
Rv0162c|M.tuberculosis_H37Rv LGSAVPSHDIPRFVSLWQSGRLPVESLVTSTIRLDDINEAMDHLADGIAV
MMAR_0405|M.marinum_M LGSAVPSRDIPRFVSLWQSGRLPVESLVSATIRLDDLNEAMDHLADGTAV
MAV_5022|M.avium_104 LGSAVPSRDIPRFVELWRAGRLPVESLVSSTIRLEDINEAMDHLADGTAV
Mflv_0725|M.gilvum_PYR-GCK EGDSIPSVFIPELLALHARGRFPIERLVT-EFSHRDVTEALAAQHAGDVV
MAB_4388|M.abscessus_ATCC_1997 EGDANPQEFIPELLQMHARGHFPIESLIT-TYPAADIEQAVADARSGAAI
TH_2558|M.thermoresistible__bu EGDAEPRTFIPRLIELWRAGRFPFDRMIE-TFPLSEINTAEQRALAGEII
MSMEG_4039|M.smegmatis_MC2_155 EGASRPHESIPRLVKMIEDGALPVHEIVT-TFPLEDIAAAIDAAKSGRVV
MUL_3861|M.ulcerans_Agy99 FGSCNPRADIARLAGLYQSGQLQLDEMITRRYRLDDINAAYTDLLNGDIV
Mvan_5270|M.vanbaalenii_PYR-1 YGEANPRADIPRLLSLYREGKLLLDETVTHEYKLAEVNEGYEDMRAGRNI
MLBr_01784|M.leprae_Br4923 YGDCLPERDFPTLVDLYLQGRLPLGKFVSERIGLGDVEEAFHKIHGGKVL
* . * :. : : * : . : :: . * :
Mb0167c|M.bovis_AF2122/97 RQLISFTGDL
Rv0162c|M.tuberculosis_H37Rv RQLISFTGDL
MMAR_0405|M.marinum_M RQIIVF----
MAV_5022|M.avium_104 RQLIRFDA--
Mflv_0725|M.gilvum_PYR-GCK KPVLTWP---
MAB_4388|M.abscessus_ATCC_1997 KPVLLW----
TH_2558|M.thermoresistible__bu KPVLIPGA--
MSMEG_4039|M.smegmatis_MC2_155 KPVLLPH---
MUL_3861|M.ulcerans_Agy99 RGIIDFTI--
Mvan_5270|M.vanbaalenii_PYR-1 RGVILHDH--
MLBr_01784|M.leprae_Br4923 RSVVML----
: ::