For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PSLATESDHSVIFVGFEGEIMKLHGATALVTGANRGLGHHLAVELIRRGAKVYATARRPELVDVPGAHVL RLDITDQWSVEAAAAAADDVDVLINNAAFTDGGNLVTGDLAKIRATMDSNYYGTLAMIRAFAPILARNGG GAILNVLSVAAWRTVDGNTSYAAAKSAEWGLTNGVRVELARQGTHVAALVPGLIRTDTLLEFARGNGIEL PEEHMNEPADLARMALDGLEAGEVEIIDRMSVDAKAALAGPPVTLSL*
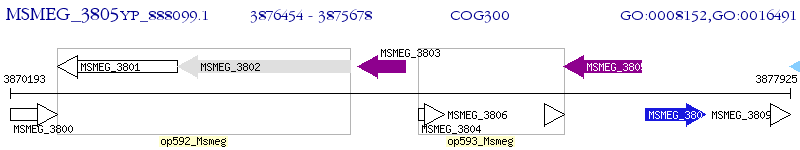
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_3805 | - | - | 100% (258) | oxidoreductase, short chain dehydrogenase/reductase family protein |
| M. smegmatis MC2 155 | MSMEG_1968 | - | 2e-55 | 52.97% (236) | putative oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_5711 | - | 1e-54 | 51.10% (227) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3199 | - | 2e-37 | 40.61% (229) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4369 | fabG | 6e-20 | 33.18% (223) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. tuberculosis H37Rv | Rv3174 | - | 5e-37 | 40.17% (229) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01094 | - | 3e-18 | 31.86% (204) | short chain alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4428 | - | 2e-36 | 40.09% (227) | short chain dehydrogenase |
| M. marinum M | MMAR_1614 | - | 5e-23 | 34.38% (224) | short chain dehydrogenase |
| M. avium 104 | MAV_4667 | - | 1e-20 | 36.57% (175) | oxidoreductase |
| M. thermoresistible (build 8) | TH_3693 | - | 8e-20 | 33.18% (220) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3423 | - | 1e-22 | 33.93% (224) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1493 | - | 1e-100 | 78.02% (232) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3805|M.smegmatis_MC2_155 MPSLATESDHSVIFVGFEGEIMKLHGATALVTGANRGLGHHLAVELIRRG
Mvan_1493|M.vanbaalenii_PYR-1 ---------------------MKLRGATALVTGANRGIGHHFAVELLQRG
Mb3199|M.bovis_AF2122/97 --------------------MTSLAERTALVTGANRGMGREYVAQLLGRK
Rv3174|M.tuberculosis_H37Rv --------------------MTSLAERTVLVTGANRGMGREYVAQLLGRK
MAB_4428|M.abscessus_ATCC_1997 ----------------------MIKDTVVLVTGGRRGLGAAIVDEVLNRG
MMAR_1614|M.marinum_M -------------------------MSTWLITGCSTGLGRALAEAVIDAG
MUL_3423|M.ulcerans_Agy99 -------------------------MSTWLITGCSTGLGRALAEAVIDAG
Mflv_4369|M.gilvum_PYR-GCK --------------------MSLLTGQTAVITGGAQGLGFAIAQRFVDEG
TH_3693|M.thermoresistible__bu --------------------VSLLAGQTAVVTGGARGLGYAIAERFIAEG
MAV_4667|M.avium_104 ------------------MVGTGRNERVAVVTGASSGIGEATARTLAAQG
MLBr_01094|M.leprae_Br4923 --------------------MEGFAGKVAVVTGAGSGIGQALAIELARSG
. ::** *:* . .
MSMEG_3805|M.smegmatis_MC2_155 -AKVYATARRPELVDVPG------AHVL--RLDITD----QWSVEAAAAA
Mvan_1493|M.vanbaalenii_PYR-1 -AKVYATARRPELVSVPG------AEVI--RLDITD----QSSVDAVAAV
Mb3199|M.bovis_AF2122/97 VAKVYAATRNPLAIDVSD------PRVIPLQLDVTD----AVSVAEAADL
Rv3174|M.tuberculosis_H37Rv VAKVYAATRNPLAIDVSD------PRVIPLQLDVTD----AVSVAEAADL
MAB_4428|M.abscessus_ATCC_1997 ARKVYSTARAP--FEDPR------AQVVTKQLEVTS----AESVDALARD
MMAR_1614|M.marinum_M HHTVATARSVGGVADLVQRS--P-ERVLPLALDITEPDQITAAVQAAQQR
MUL_3423|M.ulcerans_Agy99 HHTVATARSVGGVADLAQRS--P-ERVLPLALDITEPDQITAAMQAAQQR
Mflv_4369|M.gilvum_PYR-GCK ARVVLGDVNLEATQEAAEKLGGA-DVAHAVRCDVTSSAEVDALVAAAVDT
TH_3693|M.thermoresistible__bu ARVVIGDIDESATRAAADSLGGA-DVAVGMRCDVTRSDEVEALVGAAVKR
MAV_4667|M.avium_104 FHVVAVARRAKRIHALAN----E-IGGTAIVADVTD----DAAVAALASK
MLBr_01094|M.leprae_Br4923 AKLAISDVDGEGLAQTEGQLKAIGASARTDRLDVTEREAFLTYADVVHEN
. ::*
MSMEG_3805|M.smegmatis_MC2_155 ADDVDVLINNAAFTD-GGNLVTGDLAKIRATMDSNYYGTLAMIRAFAPIL
Mvan_1493|M.vanbaalenii_PYR-1 AGDVDVLINNAADTA-GGNLVTGDLDAIRSTMDSNYYGTLAMIRAFAPIL
Mb3199|M.bovis_AF2122/97 ATDVGILINNAGISR-ASSVLDKDTSALRGELETNLFGPLALASAFADRI
Rv3174|M.tuberculosis_H37Rv ATDVGILINNAGISR-ASSVLDKDTSALRGELETNLFGPLALASAFADRI
MAB_4428|M.abscessus_ATCC_1997 LTDVEIVVNNAGVLH-PDPLLTGDFDRIDATFQTNVFGPLRIARAFAPIL
MMAR_1614|M.marinum_M FGGIDVLVNNAGYGY-RAAVEEGDDAEVRDLFETHFFGTVALIKAVLPDM
MUL_3423|M.ulcerans_Agy99 FGGIDVLVNNAGYGY-RAAVEEGDDAEVRDLFETHFFGTVALIKAVLPDM
Mflv_4369|M.gilvum_PYR-GCK FGALDIMVNNAGITR-DATMRKMTEDDFDKVIAVHLKGTWNGLRAAAAIM
TH_3693|M.thermoresistible__bu FGSLDVMVNNAGITR-DATMRKMTEQQFDEVIAVHLKGTWNGLRAAAAIM
MAV_4667|M.avium_104 LSRVDVLVNNAGGARGLEPVAEADLEHWRWMWETNVIGTLRVTRALLPKL
MLBr_01094|M.leprae_Br4923 FGKVNQIYNNAGIAF-TGDVEVSHFKDIERVMDVDYWGVVNGTKAFLPYL
: : ***. : . * * :
MSMEG_3805|M.smegmatis_MC2_155 ARNGGGAILNVLSVAAWRTVDGNTSYAAAKSAEWGLTNGVRVELARQG--
Mvan_1493|M.vanbaalenii_PYR-1 GRNGGGAILNVLSAAAWTTVDGNTAYAAAKSAQWGLTNGVRIELAPQG--
Mb3199|M.bovis_AF2122/97 AERSG-AIVNVSSVLAWLPLG--MSYGVSKAAMWSATESMRIELAPRG--
Rv3174|M.tuberculosis_H37Rv AERSG-AIVNVSSVLAWLPLG--MSYGVSKAAMWSATESMRIELAPRG--
MAB_4428|M.abscessus_ATCC_1997 KANGGGAIVNIHSVLSWLGGAG--AYGASKAAIWSVSNTLRLELEAQH--
MMAR_1614|M.marinum_M RARRSGAIVNISSIAVALTPVGSGYYAAAKAAMEGMSGALHGELAPLG--
MUL_3423|M.ulcerans_Agy99 RARRSGAIVNISSIAVALTPVGSGYYAAAKAAMEGMSGALHGELAPLG--
Mflv_4369|M.gilvum_PYR-GCK RENKRGVIVNMSSISGKVGMIGQTNYSAAKAGIVGMTKAASKELAYLG--
TH_3693|M.thermoresistible__bu REQKRGAIVNMSSISGKVGMVGQTNYSAAKAGIVGMTKAASKELAYLG--
MAV_4667|M.avium_104 VDSGDGLIVTVTSVAALEVYDGGAGYTAAKHGQGALHRTLRGELLGKP--
MLBr_01094|M.leprae_Br4923 ISSGDGHVINISSVFGLFSVPGQAAYNSAKFAVRGFTEALREEMALAGRP
::.: * * :* . . *:
MSMEG_3805|M.smegmatis_MC2_155 THVAALVPGLIRTDTL-LEFARGNGIELPEEHMNEPADLARMALDGLEAG
Mvan_1493|M.vanbaalenii_PYR-1 TQVVALVPGLVGTQTL-FDFAERHGIEFPDGAVMDPADLVRLALDGLERG
Mb3199|M.bovis_AF2122/97 VQVVGVYVGLVDT-----DMGR-----FADAPKSDPADVVRQVLDGIEAG
Rv3174|M.tuberculosis_H37Rv VQVVGVYVGLVDT-----DMGR-----FADAPKSDPADVVRQVLDGIEAG
MAB_4428|M.abscessus_ATCC_1997 THVLGVHAAFIDT-----DMVS-----ALPVPKTPPSAIAARIIDALDKG
MMAR_1614|M.marinum_M ISVTVVEPGAFRTDFAGRSLVQSTRMINDYAATAGQRRKENDTMHGNQPG
MUL_3423|M.ulcerans_Agy99 ISVTVVEPGAFRTDFAGRSLVQSTRMINDYAATAGQRRKENDTMHGNQPG
Mflv_4369|M.gilvum_PYR-GCK VRVNAIQPGLIRS-----AMTE-----AMPQRIWDSKVAEVPLGRAGEPD
TH_3693|M.thermoresistible__bu VRVNAIQPGLIRS-----AMTE-----AMPQRIWDEKLAEIPMGRAGEPE
MAV_4667|M.avium_104 VRLTEIAPGAVETEFSLVRFDG-------DQQRADAVYTGITPLVAADVA
MLBr_01094|M.leprae_Br4923 VNVTTVYPGGIKTAIARNATAAEG----LDVSKIASRFDTWVAHTSPQHA
: : . . : . :
MSMEG_3805|M.smegmatis_MC2_155 -EVEIIDRMSVDAKA------ALAGPPVTLSL------------------
Mvan_1493|M.vanbaalenii_PYR-1 -EIEILDPMGVDAKT------SLAGPPRAFAPSAFDAG------------
Mb3199|M.bovis_AF2122/97 KEDVLADEMSRQVRA------SLNVPARERIARLMGN-------------
Rv3174|M.tuberculosis_H37Rv KEDVLADEMSRQVRA------SLNVPARERIARLMGN-------------
MAB_4428|M.abscessus_ATCC_1997 DNEVLTDDLTVQIKS------QLAGPVEKLAYRPGH--------------
MMAR_1614|M.marinum_M DPAKAAAAIITAVESPAPPAFLLLGPDALMAYRHIAQRRDVEITDWEQLT
MUL_3423|M.ulcerans_Agy99 DPAKAAAAIITAVESPAPPAFLLLGPDALMAYRHIAQRRDVEITDWEQLT
Mflv_4369|M.gilvum_PYR-GCK EVAKVALFLASDLSS------YMTGTVLEVTGGRHL--------------
TH_3693|M.thermoresistible__bu EVAKVALFLASDLSS------YMTGTVLEVTGGRHL--------------
MAV_4667|M.avium_104 EVIGFVASRPAHVNLD----LIVMKPRDQASASRFNRRG-----------
MLBr_01094|M.leprae_Br4923 ARIILKAVRKKKARVLVGPDAKVANVVVRFSGGAGYQRLFAQVASRLILN
:
MSMEG_3805|M.smegmatis_MC2_155 -------
Mvan_1493|M.vanbaalenii_PYR-1 -------
Mb3199|M.bovis_AF2122/97 -------
Rv3174|M.tuberculosis_H37Rv -------
MAB_4428|M.abscessus_ATCC_1997 -------
MMAR_1614|M.marinum_M AGTNLTP
MUL_3423|M.ulcerans_Agy99 AGTNLTP
Mflv_4369|M.gilvum_PYR-GCK -------
TH_3693|M.thermoresistible__bu -------
MAV_4667|M.avium_104 -------
MLBr_01094|M.leprae_Br4923 QR-----