For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DIDGTTAVVTGGGSGIGAALAEAFAAAGARVVVADLDEPGAAATAARIESAGGNAVAVRADASATADIEA LTRTEFGPVDIYVANAGIIGASGLGTDEEWDRILAVNLRAHIRAADVLVPQWISRGRGYFVSVASAAGLL SQIGAAGYAVTKHAAVGFAEWLAITYGDDGVGVSCVCPLGVDTPLLHAVRNSAETDALVGAESIVQSGDV ITPQDVATVTVEAVQAGRFLVLPHPHVLDLYRRKGSDYDRWIAGMRRYQRSLRGTAVPGT*
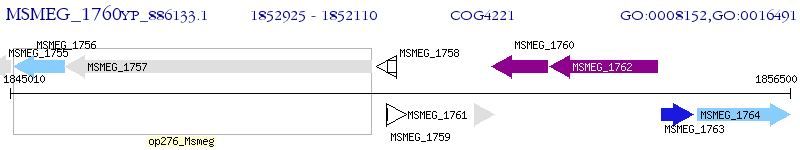
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1760 | - | - | 100% (271) | short-chain dehydrogenase/reductase SDR |
| M. smegmatis MC2 155 | MSMEG_5235 | - | 1e-29 | 34.02% (241) | short chain dehydrogenase family protein |
| M. smegmatis MC2 155 | MSMEG_0527 | - | 3e-25 | 33.94% (218) | 2-hydroxycyclohexnecarboxyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0874c | - | 1e-30 | 33.33% (261) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4800 | - | 2e-99 | 67.42% (267) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0851c | - | 3e-31 | 33.72% (261) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 3e-25 | 38.42% (203) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3103c | - | 5e-85 | 60.30% (267) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_4755 | - | 2e-31 | 30.92% (262) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_0895 | - | 1e-31 | 32.69% (260) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3454 | - | 1e-101 | 69.17% (266) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_0326 | - | 1e-30 | 30.53% (262) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_1642 | - | 1e-102 | 68.54% (267) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0895|M.avium_104 --------------------------------------------------
MMAR_4755|M.marinum_M --------------------------------------------------
MUL_0326|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4800|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1642|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1760|M.smegmatis_MC2_155 --------------------------------------------------
TH_3454|M.thermoresistible__bu --------------------------------------------------
MAB_3103c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0895|M.avium_104 --------------------------------------------------
MMAR_4755|M.marinum_M --------------------------------------------------
MUL_0326|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4800|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1642|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1760|M.smegmatis_MC2_155 --------------------------------------------------
TH_3454|M.thermoresistible__bu --------------------------------------------------
MAB_3103c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0895|M.avium_104 --------------------------------------------------
MMAR_4755|M.marinum_M --------------------------------------------------
MUL_0326|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4800|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1642|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1760|M.smegmatis_MC2_155 --------------------------------------------------
TH_3454|M.thermoresistible__bu --------------------------------------------------
MAB_3103c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0895|M.avium_104 --------------------------------------------------
MMAR_4755|M.marinum_M --------------------------------------------------
MUL_0326|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4800|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1642|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1760|M.smegmatis_MC2_155 --------------------------------------------------
TH_3454|M.thermoresistible__bu --------------------------------------------------
MAB_3103c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0895|M.avium_104 --------------------------------------------------
MMAR_4755|M.marinum_M --------------------------------------------------
MUL_0326|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4800|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1642|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1760|M.smegmatis_MC2_155 --------------------------------------------------
TH_3454|M.thermoresistible__bu --------------------------------------------------
MAB_3103c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
Mb0874c|M.bovis_AF2122/97 --------------------------------------------------
Rv0851c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0895|M.avium_104 --------------------------------------------------
MMAR_4755|M.marinum_M --------------------------------------------------
MUL_0326|M.ulcerans_Agy99 --------------------------------------------------
Mflv_4800|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1642|M.vanbaalenii_PYR-1 ---------------------------------------MRERACLRPTR
MSMEG_1760|M.smegmatis_MC2_155 --------------------------------------------------
TH_3454|M.thermoresistible__bu --------------------------------------------------
MAB_3103c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
Mb0874c|M.bovis_AF2122/97 ------------------MDGFP----GRGAVITGGASGIGLATGTEFAR
Rv0851c|M.tuberculosis_H37Rv ------------------MDGFP----GRGAVITGGASGIGLATGTEFAR
MAV_0895|M.avium_104 ------------------MDGFLSGFDGRAAVVTGGASGIGLATATEFAR
MMAR_4755|M.marinum_M ------------------MDGFA----GRGAVITGGASGIGLATATEFAR
MUL_0326|M.ulcerans_Agy99 ------------------MDGFA----GSGAVITGGASGIGLATATEFAR
Mflv_4800|M.gilvum_PYR-GCK ---------------MELAG--------RTAIVTGAASGIGAALATELAR
Mvan_1642|M.vanbaalenii_PYR-1 RLWDELCARSRHTLGMELAR--------KTAIVTGGASGIGAAIASELAA
MSMEG_1760|M.smegmatis_MC2_155 ---------------MDIDG--------TTAVVTGGGSGIGAALAEAFAA
TH_3454|M.thermoresistible__bu ---------------MRIDN--------SVFVVTGAGSGIGRALALSFAD
MAB_3103c|M.abscessus_ATCC_199 ---------------MDING--------SVAIVTGAGSGIGQALAWGLVD
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
:**..**** . :.
Mb0874c|M.bovis_AF2122/97 RGARVVLGDVDKPGLRQAVNHLRAEGFDVHGVMCDVRHREEVTHLADEAF
Rv0851c|M.tuberculosis_H37Rv RGARVVLGDVDKPGLRQAVNHLRAEGFDVHSVMCDVRHREEVTHLADEAF
MAV_0895|M.avium_104 RGARLVLSDVDQPALEQAVNGLRGQGFDAHGVVCDVRHLDEMVRLADEAF
MMAR_4755|M.marinum_M RGAKVVLADIDKPGLERSVEHLRGKGFDAHGVMCDVRHLSEVNHLAAESA
MUL_0326|M.ulcerans_Agy99 RGAKVVLADIDKPGLERSVEHLRGKGFDAHGVMCDVRHLSEVNHLAAESA
Mflv_4800|M.gilvum_PYR-GCK RGVSVVIGDLDDDGAHATASAIRADGGAAAALRADATSVADIEALITLAS
Mvan_1642|M.vanbaalenii_PYR-1 HGVSVVVGDLDEAGAHTTAAAIRDSGGAATALRADASDVVDIEGLITLAG
MSMEG_1760|M.smegmatis_MC2_155 AGARVVVADLDEPGAAATAARIESAGGNAVAVRADASATADIEALT---R
TH_3454|M.thermoresistible__bu AGARVVAGDIDESAADTTAATIRDSGGHAVPIRADATTPAGIDAMISLAQ
MAB_3103c|M.abscessus_ATCC_199 AGATVVASDIDADAVGRTAAARDG----IVGQRADASAVSDIEDLIALAE
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFAEQVS
*. :* .*:* . :. *. : :
Mb0874c|M.bovis_AF2122/97 RLLGHFDVVFSNAGIVVGGPIVEMTHDDWRWVIDVDLWGSIHTVEAFLPR
Rv0851c|M.tuberculosis_H37Rv RLLGHVDVVFSNAGIVVGGPIVEMTHDDWRWVIDVDLWGSIHTVEAFLPR
MAV_0895|M.avium_104 RLLGGVDVVFSNAGIVVAGPLAQMNHDDWRWVIDIDLWGSIHAVEAFLPR
MMAR_4755|M.marinum_M RLLGQIDVVFSNAGIVVAGPIADMTHDDWRWVIDIDLWGSIHTVEAFLPK
MUL_0326|M.ulcerans_Agy99 RLLGQIDVVFSNAGIVVAGPIADMTHDVWRWVIDIDLWGSIHTVEAFLPK
Mflv_4800|M.gilvum_PYR-GCK REFTPVDIYVANAGVIGKPGLGT--ESDWDVILDVNLRAHVRAARLLVPH
Mvan_1642|M.vanbaalenii_PYR-1 REFAPVDIYVANAGITGKPGLGT--ESDWDVVLDVNLRAHVRAARLVVPH
MSMEG_1760|M.smegmatis_MC2_155 TEFGPVDIYVANAGIIGASGLGT--DEEWDRILAVNLRAHIRAADVLVPQ
TH_3454|M.thermoresistible__bu ETFGPVDGYVANAGVIGPPGLGT--DTGWDRVLDVNLRAHVRAADALVPQ
MAB_3103c|M.abscessus_ATCC_199 KEFGPVDLYAANAGIAGGLGLGIG-EAAWDLTIDVNLRAHIRAATLLVPG
MLBr_00862|M.leprae_Br4923 AKHGVPDIVVNNAGIGQAGRFLDTPPEEFDRVLAVNLGGVVNCCRAFGQR
* ***: : : : ::* . :. .
Mb0874c|M.bovis_AF2122/97 LLEQGTGGHVVFTASFAGLVPNAGLGAYGVAKYGVVGLAETLAREVTADG
Rv0851c|M.tuberculosis_H37Rv LLEQGTGGHVVFTASFAGLVPNAGLGAYGVAKYGVVGLAETLAREVTADG
MAV_0895|M.avium_104 LLEQGTGGHIAFTASFAGLVPNAGLGTYGVAKYGVVGLAETLAREVKPNG
MMAR_4755|M.marinum_M LLEHG--GHIAFTASFAGLVPNAGLGAYGVAKYGVVGLAETLSREMKDRG
MUL_0326|M.ulcerans_Agy99 LLEHG--GHIAFTASFAGLVPNAGLGAYGVAKYGVVGLAETLSREMKDRG
Mflv_4800|M.gilvum_PYR-GCK WQSVGS-GYFVSVASAAGLLTQLGAAGYAVSKHAAVGFAEWLAVTYGDDG
Mvan_1642|M.vanbaalenii_PYR-1 WQSRGS-GYFVSVASAAGLLTQLGAAGYAVTKHAAVGFAEWLAITYGDDG
MSMEG_1760|M.smegmatis_MC2_155 WISRGR-GYFVSVASAAGLLSQIGAAGYAVTKHAAVGFAEWLAITYGDDG
TH_3454|M.thermoresistible__bu WISRGT-GYFVSVASAAGLLTQLGAAAYAVSKHAAVGFAEWLAITYGDRG
MAB_3103c|M.abscessus_ATCC_199 WVERGR-GYFLSVASAAGLLTQIGSPAYSVTKHAAVGFAEWLSITYGGQG
MLBr_00862|M.leprae_Br4923 LVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRAELDAVG
. * *:. .:* *. . . * .:* .. ::: * *
Mb0874c|M.bovis_AF2122/97 IGVSVLCPMVVETNLVANSERIRGAACAQSST---TGSPGPLPLQDDNLG
Rv0851c|M.tuberculosis_H37Rv IGVSVLCPMVVETNLVANSERIRGAACAQSST---TGSPGPLPLQDDNLG
MAV_0895|M.avium_104 IGVSVLCPMVVETKLVSNSERIRGADYGMSATP--EGAFGPLPTQDESVS
MMAR_4755|M.marinum_M VGVTVLCPMVVETRLVSNSERIRGADYGLSSAPQVTGDMGSLPAQDDSLS
MUL_0326|M.ulcerans_Agy99 VGVTVLCPMVVETQLVSNSERIRGADYGLSSAPQVTGDMGSLPAQDDSLS
Mflv_4800|M.gilvum_PYR-GCK IGVSCVCPLGVKTPLLQGIRGIDDPDARLGAS--------SIEQSAGVIS
Mvan_1642|M.vanbaalenii_PYR-1 IGVSCVCPLGVDTPLLQAIRSTDDPDSRLGAS--------SIEQSGSVIS
MSMEG_1760|M.smegmatis_MC2_155 VGVSCVCPLGVDTPLLHAVRNSAETDALVGAE--------SIVQSGDVIT
TH_3454|M.thermoresistible__bu IGVSCVCPMGVRTPLLGAVRDSTDTTAQLAAR--------SITSAGAVLD
MAB_3103c|M.abscessus_ATCC_199 VGVSCLCPMGVKTALLDGLTESDDPDVRVAGT--------SITAAGDVLE
MLBr_00862|M.leprae_Br4923 VGLTTICPGVIDTNIIQSTRIDTPTAKQERVRD--RRGQLDMMFRLRRYG
:*:: :** : * :: . :
Mb0874c|M.bovis_AF2122/97 VDDIAQLTADAILANRLYVLPHAASRASIRRRFERIDRTFDEQAAEGWRH
Rv0851c|M.tuberculosis_H37Rv VDDIAQLTADAILANRLYVLPHAASRASIRRRFERIDRTFDEQAAEGWRH
MAV_0895|M.avium_104 ADDVARLTADAILANRLYILPHAAARESIRRRFERIDRTFDEQAAEGWTH
MMAR_4755|M.marinum_M VDELARLTADAILANRLYVLPHEGARTSIQRRFARIDRTFDEQAAEGWRH
MUL_0326|M.ulcerans_Agy99 VDELARLTADAILANGLYVLPHEGARTSIQRRFARIDRTFDEQAAEGWRH
Mflv_4800|M.gilvum_PYR-GCK AADVATVTVEAIREERFLVLPHAELLEMYRRKGGDYDRWISGMRRYQRTL
Mvan_1642|M.vanbaalenii_PYR-1 AADVAAVTVEAIRTDRFLVLPHPELLEMYRNKGGDYDRWIGGMRRYQRTL
MSMEG_1760|M.smegmatis_MC2_155 PQDVATVTVEAVQAGRFLVLPHPHVLDLYRRKGSDYDRWIAGMRRYQRSL
TH_3454|M.thermoresistible__bu AAEVADLTVAAVREGRFLVLPHPEVLDMYRQKGADYDRWIAGMRRYQRSL
MAB_3103c|M.abscessus_ATCC_199 PAVVAAMALDAIRDEKFLVLPHPQVLDMYRQKGADYDRWIAGMRRYQAIL
MLBr_00862|M.leprae_Br4923 PDKVADTILSAIKKNKPIRPVAPEAYALYG-----ISRVLPQVLRSTARI
:* *: .* :
Mb0874c|M.bovis_AF2122/97 --------
Rv0851c|M.tuberculosis_H37Rv --------
MAV_0895|M.avium_104 --------
MMAR_4755|M.marinum_M --------
MUL_0326|M.ulcerans_Agy99 --------
Mflv_4800|M.gilvum_PYR-GCK RSL-----
Mvan_1642|M.vanbaalenii_PYR-1 RAL-----
MSMEG_1760|M.smegmatis_MC2_155 RGTAVPGT
TH_3454|M.thermoresistible__bu AAER----
MAB_3103c|M.abscessus_ATCC_199 EDQIIR--
MLBr_00862|M.leprae_Br4923 RVI-----