For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDGFAGSGAVITGGASGIGLATATEFARRGAKVVLADIDKPGLERSVEHLRGKGFDAHGVMCDVRHLSEV NHLAAESARLLGQIDVVFSNAGIVVAGPIADMTHDVWRWVIDIDLWGSIHTVEAFLPKLLEHGGHIAFTA SFAGLVPNAGLGAYGVAKYGVVGLAETLSREMKDRGVGVTVLCPMVVETQLVSNSERIRGADYGLSSAPQ VTGDMGSLPAQDDSLSVDELARLTADAILANGLYVLPHEGARTSIQRRFARIDRTFDEQAAEGWRH
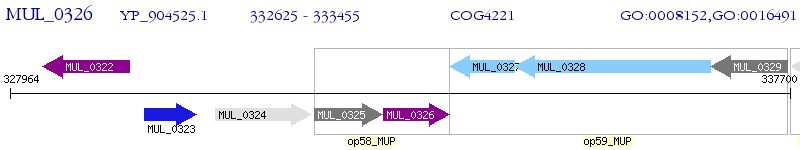
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0326 | - | - | 100% (276) | short chain dehydrogenase |
| M. ulcerans Agy99 | MUL_3643 | - | 4e-32 | 35.86% (251) | short-chain membrane-associated dehydrogenase |
| M. ulcerans Agy99 | MUL_3160 | - | 8e-31 | 35.68% (199) | short-chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0874c | - | 1e-120 | 76.26% (278) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_3370 | - | 2e-32 | 39.47% (190) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0851c | - | 1e-119 | 75.90% (278) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_01094 | - | 5e-28 | 36.23% (207) | short chain alcohol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4478c | - | 3e-33 | 38.89% (252) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_4755 | - | 1e-155 | 98.55% (276) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_0895 | - | 1e-120 | 75.90% (278) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2291 | - | 9e-31 | 37.93% (203) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_2445 | - | 2e-28 | 35.81% (229) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_0852 | - | 1e-32 | 37.10% (248) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2291|M.smegmatis_MC2_155 --MAQGSDFA-------GKRCFVTGAASGIGRATALALAAAGAELYLTDR
Mflv_3370|M.gilvum_PYR-GCK MASTDTAQLS-------GKVAFVTGGASGIGAALVAKLATAGTEVWIADR
TH_2445|M.thermoresistible__bu ------MTFA-------GKHALVTGAGSGIGAALCRALAAAGAEVMCTDI
Mvan_0852|M.vanbaalenii_PYR-1 ---MVSNHFR-------GKVCVVTGAASGIGLAVGAALLDHGATVVLADF
MUL_0326|M.ulcerans_Agy99 -----MDGFA-------GSGAVITGGASGIGLATATEFARRGAKVVLADI
MMAR_4755|M.marinum_M -----MDGFA-------GRGAVITGGASGIGLATATEFARRGAKVVLADI
Mb0874c|M.bovis_AF2122/97 -----MDGFP-------GRGAVITGGASGIGLATGTEFARRGARVVLGDV
Rv0851c|M.tuberculosis_H37Rv -----MDGFP-------GRGAVITGGASGIGLATGTEFARRGARVVLGDV
MAV_0895|M.avium_104 -----MDGFLSG---FDGRAAVVTGGASGIGLATATEFARRGARLVLSDV
MAB_4478c|M.abscessus_ATCC_199 --MVSLLGFGRGERRTRKVDAVVTGAASGIGRAFAVELARRGGRVVCADV
MLBr_01094|M.leprae_Br4923 -----MEGFA-------GKVAVVTGAGSGIGQALAIELARSGAKLAISDV
: ..:**..**** * : * : *
MSMEG_2291|M.smegmatis_MC2_155 DADGLVQTVADARALGAEVPAHRALDISDYDQVREFAKDIHAAHG-SMDV
Mflv_3370|M.gilvum_PYR-GCK QIAPAEELAAQLRATGAVA-HAVELDVRDPAAFAHAVHTAVDTSG-RLDY
TH_2445|M.thermoresistible__bu DGAAAERTAQSIGGR------WAALDVTDAAAVQAAVDGVVDRAG-RLDL
Mvan_0852|M.vanbaalenii_PYR-1 DADRLTTVAEGLCMHDGRV-HSAVVDVTVQEAVQALVEGAARAHG-SLDF
MUL_0326|M.ulcerans_Agy99 DKPGLERSVEHLRGKG-FDAHGVMCDVRHLSEVNHLAAESARLLG-QIDV
MMAR_4755|M.marinum_M DKPGLERSVEHLRGKG-FDAHGVMCDVRHLSEVNHLAAESARLLG-QIDV
Mb0874c|M.bovis_AF2122/97 DKPGLRQAVNHLRAEG-FDVHGVMCDVRHREEVTHLADEAFRLLG-HFDV
Rv0851c|M.tuberculosis_H37Rv DKPGLRQAVNHLRAEG-FDVHSVMCDVRHREEVTHLADEAFRLLG-HVDV
MAV_0895|M.avium_104 DQPALEQAVNGLRGQG-FDAHGVVCDVRHLDEMVRLADEAFRLLG-GVDV
MAB_4478c|M.abscessus_ATCC_199 DLPGAKETAALIGERA-IEAA---CDVSVFDEVEALADTAEEWFGKPSDL
MLBr_01094|M.leprae_Br4923 DGEGLAQTEGQLKAIG-ASARTDRLDVTEREAFLTYADVVHENFG-KVNQ
: *: . . * :
MSMEG_2291|M.smegmatis_MC2_155 VMNIAGVSAWGTVDRLS-HQQWRSMVDINLMGPIHVIEEFIPPMIEARRG
Mflv_3370|M.gilvum_PYR-GCK LFNNAGIGIGGEVDTLT-LDDWTDIIDVNLKGVIHGVHAAYPQMIRQGSG
TH_2445|M.thermoresistible__bu LFNNAGIVWGGDTELLT-LDQWNAIIDVNIRGVVHGVAAAYPLMVRQGHG
Mvan_0852|M.vanbaalenii_PYR-1 LFNNAGVGGTMPIGVAT-LEQWRRIVDLNLWGVIYGVHAAVPIMVRQGSG
MUL_0326|M.ulcerans_Agy99 VFSNAGIVVAGPIADMT-HDVWRWVIDIDLWGSIHTVEAFLPKLLEHG--
MMAR_4755|M.marinum_M VFSNAGIVVAGPIADMT-HDDWRWVIDIDLWGSIHTVEAFLPKLLEHG--
Mb0874c|M.bovis_AF2122/97 VFSNAGIVVGGPIVEMT-HDDWRWVIDVDLWGSIHTVEAFLPRLLEQGTG
Rv0851c|M.tuberculosis_H37Rv VFSNAGIVVGGPIVEMT-HDDWRWVIDVDLWGSIHTVEAFLPRLLEQGTG
MAV_0895|M.avium_104 VFSNAGIVVAGPLAQMN-HDDWRWVIDIDLWGSIHAVEAFLPRLLEQGTG
MAB_4478c|M.abscessus_ATCC_199 VINNAGIGAGGQVVGAAPLADWQKTLGVNLWGVIHGSHVFAPRLRALGR-
MLBr_01094|M.leprae_Br4923 IYNNAGIAFTGDVEVSH-FKDIERVMDVDYWGVVNGTKAFLPYLISSGDG
: . **: :.:: * : * :
MSMEG_2291|M.smegmatis_MC2_155 GHLVNVSSAAGLVALPWHGAYSASKFGLRGVSEVLRFDLARHR--IGVSV
Mflv_3370|M.gilvum_PYR-GCK -HIVNTASMAGLITTSAQGGYSATKHAVVALSKTMRVEAATHG--VRVSV
TH_2445|M.thermoresistible__bu -HIVNTASMAGLAAAGQLTSYVMTKHAVVGLSLALRSEAAAHG--VGVLA
Mvan_0852|M.vanbaalenii_PYR-1 -HIVNTASLAGLVPFPYQSLYCTTKYGVVGLSESLRYELAADG--VRVSV
MUL_0326|M.ulcerans_Agy99 GHIAFTASFAGLVPNAGLGAYGVAKYGVVGLAETLSREMKDRG--VGVTV
MMAR_4755|M.marinum_M GHIAFTASFAGLVPNAGLGAYGVAKYGVVGLAETLSREMKDRG--VGVTV
Mb0874c|M.bovis_AF2122/97 GHVVFTASFAGLVPNAGLGAYGVAKYGVVGLAETLAREVTADG--IGVSV
Rv0851c|M.tuberculosis_H37Rv GHVVFTASFAGLVPNAGLGAYGVAKYGVVGLAETLAREVTADG--IGVSV
MAV_0895|M.avium_104 GHIAFTASFAGLVPNAGLGTYGVAKYGVVGLAETLAREVKPNG--IGVSV
MAB_4478c|M.abscessus_ATCC_199 GGIINVASAASFAAAPRMGAYNVSKAGVLALSETLAAELSGTG--VAVTV
MLBr_01094|M.leprae_Br4923 -HVINISSVFGLFSVPGQAAYNSAKFAVRGFTEALREEMALAGRPVNVTT
: :* .: . * :* .: ..: : : : * .
MSMEG_2291|M.smegmatis_MC2_155 VVPGAVKTGLVQTVEIAGVDREDPDVKK------WVDRFAGHAISPEKAA
Mflv_3370|M.gilvum_PYR-GCK LCPGVVRTPILTGGAFGRNKSVSREDQL------KLGEALR-PMDAGVFA
TH_2445|M.thermoresistible__bu VCPAAVETPLLDKGAVG---GFLGRDYF------IRGQGVRSAYDPDRLA
Mvan_0852|M.vanbaalenii_PYR-1 VCPGNVVSRIFGTPILGRPVDAAPPEDS------IPAEEAARLILAGVAA
MUL_0326|M.ulcerans_Agy99 LCPMVVETQLVSNSERIRGADYGLSSAPQVTGDMGSLPAQDDSLSVDELA
MMAR_4755|M.marinum_M LCPMVVETRLVSNSERIRGADYGLSSAPQVTGDMGSLPAQDDSLSVDELA
Mb0874c|M.bovis_AF2122/97 LCPMVVETNLVANSERIRGAACAQSST---TGSPGPLPLQDDNLGVDDIA
Rv0851c|M.tuberculosis_H37Rv LCPMVVETNLVANSERIRGAACAQSST---TGSPGPLPLQDDNLGVDDIA
MAV_0895|M.avium_104 LCPMVVETKLVSNSERIRGADYGMSATP--EGAFGPLPTQDESVSADDVA
MAB_4478c|M.abscessus_ATCC_199 LCPTFVKTSIVDSG-IIEESAARLAAT----------AMKWTGVSADSVA
MLBr_01094|M.leprae_Br4923 VYPGGIKTAIARNATAAEGLDVSKIASR--------FDTWVAHTSPQHAA
: * : : : *
MSMEG_2291|M.smegmatis_MC2_155 AKILAGVRRNRFLIYTSADIRALYTFKRVAWWPYSVAMRQVNVLFTRALR
Mflv_3370|M.gilvum_PYR-GCK QKALRGVLRNEAIIVVPRWWKALWYLERLSPALSMRTAAVALKRTREVQS
TH_2445|M.thermoresistible__bu SDTLRAVQRNKALLVKPRVAHAGWLFARLAPGLMQRASIRFVERQRAGQR
Mvan_0852|M.vanbaalenii_PYR-1 GEGIIAFPEKPRELWRSYAESPDSVEGYLKEVARQRRAAFATGDAQALFR
MUL_0326|M.ulcerans_Agy99 RLTADAILANGLYVLPHEGARTSIQRR-FARIDRTFDEQAAEGWRH----
MMAR_4755|M.marinum_M RLTADAILANRLYVLPHEGARTSIQRR-FARIDRTFDEQAAEGWRH----
Mb0874c|M.bovis_AF2122/97 QLTADAILANRLYVLPHAASRASIRRR-FERIDRTFDEQAAEGWRH----
Rv0851c|M.tuberculosis_H37Rv QLTADAILANRLYVLPHAASRASIRRR-FERIDRTFDEQAAEGWRH----
MAV_0895|M.avium_104 RLTADAILANRLYILPHAAARESIRRR-FERIDRTFDEQAAEGWTH----
MAB_4478c|M.abscessus_ATCC_199 RGCLDAHDRGQLYVLPQFDARFMWQAKRLAPDTYTKALGIAERFSRQRGN
MLBr_01094|M.leprae_Br4923 RIILKAVRKKKARVLVGPDAKVANVVVRFSGGAGYQRLFAQVASRLILNQ
. : .
MSMEG_2291|M.smegmatis_MC2_155 SGTSPR--
Mflv_3370|M.gilvum_PYR-GCK AVPLQ---
TH_2445|M.thermoresistible__bu AAQVGR--
Mvan_0852|M.vanbaalenii_PYR-1 ATQSAGGT
MUL_0326|M.ulcerans_Agy99 --------
MMAR_4755|M.marinum_M --------
Mb0874c|M.bovis_AF2122/97 --------
Rv0851c|M.tuberculosis_H37Rv --------
MAV_0895|M.avium_104 --------
MAB_4478c|M.abscessus_ATCC_199 --------
MLBr_01094|M.leprae_Br4923 R-------