For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TQRLDGRKVLITGANGGLGEEFVRQALARGAARVYATARRPHPWEDARVVPLPLDLTDPQSITTAAAAAR DVDLVINNAAIAPSDDTSVLTQDDEITRRVFETNFFGTLRVARAFAPVLADNGGGAFLNVLSAAIWIPVP TAYAASKAAAWSATNAMRAELTAQGTTVTAVVVGMVDTAMSARWDMPKVSAESVVEQSLDGVLSGAFEVL ADDDSRLAKSLLSEPYEELHAVAVQALSDFKP*
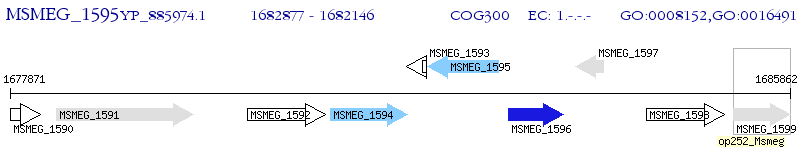
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1595 | - | - | 100% (243) | putative oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_1968 | - | 4e-52 | 50.22% (227) | putative oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_2899 | - | 5e-48 | 46.88% (224) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3199 | - | 2e-45 | 45.98% (224) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0930 | - | 2e-18 | 35.96% (178) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv3174 | - | 6e-46 | 46.43% (224) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00862 | ephD | 3e-15 | 30.17% (232) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4428 | - | 3e-41 | 42.08% (221) | short chain dehydrogenase |
| M. marinum M | MMAR_0254 | - | 9e-19 | 30.17% (242) | ketoacyl reductase |
| M. avium 104 | MAV_4667 | - | 4e-16 | 29.78% (225) | oxidoreductase |
| M. thermoresistible (build 8) | TH_3037 | - | 1e-16 | 29.33% (208) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3423 | - | 1e-18 | 36.02% (186) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0234 | - | 6e-44 | 47.47% (217) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0930|M.gilvum_PYR-GCK --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1595|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
TH_3037|M.thermoresistible__bu --------------------------------------------------
MMAR_0254|M.marinum_M --------------------------------------------------
MAV_4667|M.avium_104 --------------------------------------------------
Mflv_0930|M.gilvum_PYR-GCK --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1595|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
TH_3037|M.thermoresistible__bu --------------------------------------------------
MMAR_0254|M.marinum_M --------------------------------------------------
MAV_4667|M.avium_104 --------------------------------------------------
Mflv_0930|M.gilvum_PYR-GCK --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1595|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
TH_3037|M.thermoresistible__bu --------------------------------------------------
MMAR_0254|M.marinum_M --------------------------------------------------
MAV_4667|M.avium_104 --------------------------------------------------
Mflv_0930|M.gilvum_PYR-GCK --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1595|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
TH_3037|M.thermoresistible__bu --------------------------------------------------
MMAR_0254|M.marinum_M --------------------------------------------------
MAV_4667|M.avium_104 --------------------------------------------------
Mflv_0930|M.gilvum_PYR-GCK --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1595|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
TH_3037|M.thermoresistible__bu --------------------------------------------------
MMAR_0254|M.marinum_M --------------------------------------------------
MAV_4667|M.avium_104 --------------------------------------------------
Mflv_0930|M.gilvum_PYR-GCK --------------------------------------------------
MUL_3423|M.ulcerans_Agy99 --------------------------------------------------
Mb3199|M.bovis_AF2122/97 --------------------------------------------------
Rv3174|M.tuberculosis_H37Rv --------------------------------------------------
Mvan_0234|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1595|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4428|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
TH_3037|M.thermoresistible__bu --------------------------------------------------
MMAR_0254|M.marinum_M --------------------------------------------------
MAV_4667|M.avium_104 --------------------------------------------------
Mflv_0930|M.gilvum_PYR-GCK --------------------------MSKRWFVTGGTPGGFGVAFAEAAL
MUL_3423|M.ulcerans_Agy99 --------------------------MST-WLITGCSTG-LGRALAEAVI
Mb3199|M.bovis_AF2122/97 ---------------------MTS-LAERTALVTGANRG-MGREYVAQLL
Rv3174|M.tuberculosis_H37Rv ---------------------MTS-LAERTVLVTGANRG-MGREYVAQLL
Mvan_0234|M.vanbaalenii_PYR-1 ---------------------MTD-TEHTTALVTGANRG-LGRRFAEALV
MSMEG_1595|M.smegmatis_MC2_155 ---------------------MTQRLDGRKVLITGANGG-LGEEFVRQAL
MAB_4428|M.abscessus_ATCC_1997 ---------------------MIK---DTVVLVTGGRRG-LGAAIVDEVL
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSG-IGRETALALA
TH_3037|M.thermoresistible__bu ----------------------MGQFDDKVAIVTGAGGG-IGQAYAEALA
MMAR_0254|M.marinum_M -------------------MALPPPGKDRAAIVTGASSG-IGEEFARILS
MAV_4667|M.avium_104 --------------------MVGTGRNERVAVVTGASSG-IGEATARTLA
:** * :* .
Mflv_0930|M.gilvum_PYR-GCK EAGDRVVITSRRPHELAAWADGYD---DRVLVVPLELTDAAQVQRVVQAA
MUL_3423|M.ulcerans_Agy99 DAGHHTVATARSVGGVADLAQRSP---ERVLPLALDITEPDQITAAMQAA
Mb3199|M.bovis_AF2122/97 GRKVAKVYAATRNPLAIDVSD------PRVIPLQLDVTDAVSVAEAADLA
Rv3174|M.tuberculosis_H37Rv GRKVAKVYAATRNPLAIDVSD------PRVIPLQLDVTDAVSVAEAADLA
Mvan_0234|M.vanbaalenii_PYR-1 ARG-AKVYAAARRPETIDI--------PGVVPIQLDITDPESVRRAAEQA
MSMEG_1595|M.smegmatis_MC2_155 ARGAARVYATARRPHPWED--------ARVVPLPLDLTDPQSITTAAAAA
MAB_4428|M.abscessus_ATCC_1997 NRGARKVYSTARAP--FEDPR------AQVVTKQLEVTSAESVDALARDL
MLBr_00862|M.leprae_Br4923 RRGAEVVVSDIDEAAAKNTAAQIVASGGVAYAYALDVSDAAAVEAFAEQV
TH_3037|M.thermoresistible__bu REGAAVVVADINTEGAQKVADGIKGEGGDALAVPVDVSDPESAEEMAAQT
MMAR_0254|M.marinum_M QRGYQVVLVARSADRLEALAGRLG---SDTHPLPADLSVRSDRAGLVDRV
MAV_4667|M.avium_104 AQGFHVVAVARRAKRIHALANEIG-----GTAIVADVTDDAAVAALASKL
* :::
Mflv_0930|M.gilvum_PYR-GCK EEHFGGIDVLVNNAGRG---WYGSIEGMDESSLRAMFELNFFAVLSVTRA
MUL_3423|M.ulcerans_Agy99 QQRFGGIDVLVNNAGYG---YRAAVEEGDDAEVRDLFETHFFGTVALIKA
Mb3199|M.bovis_AF2122/97 TD----VGILINNAGIS---RASSVLDKDTSALRGELETNLFGPLALASA
Rv3174|M.tuberculosis_H37Rv TD----VGILINNAGIS---RASSVLDKDTSALRGELETNLFGPLALASA
Mvan_0234|M.vanbaalenii_PYR-1 TD----VTVVINNAGVS---TRAGLLDGTLEDIRLEMDTHYFGTLAVTRA
MSMEG_1595|M.smegmatis_MC2_155 RD----VDLVINNAAIAP-SDDTSVLTQDDEITRRVFETNFFGTLRVARA
MAB_4428|M.abscessus_ATCC_1997 TD----VEIVVNNAGVL---HPDPLLTGDFDRIDATFQTNVFGPLRIARA
MLBr_00862|M.leprae_Br4923 SAKHGVPDIVVNNAGIG---QAGRFLDTPPEEFDRVLAVNLGGVVNCCRA
TH_3037|M.thermoresistible__bu VSEFGGIDYLVNNAAIFGGMKLDFLITVDWDYYKRFMSVNLDGALVCTRA
MMAR_0254|M.marinum_M AALGLVPDILINNAGLS---TLGPVAKSVPEQELNLVEVDVAAVVDLCSR
MAV_4667|M.avium_104 SR----VDVLVNNAGGAR--GLEPVAEADLEHWRWMWETNVIGTLRVTRA
::***. . . . :
Mflv_0930|M.gilvum_PYR-GCK VLPGMRARGSG-WIVNVSSVAGLVAAPGFGYYSATKFAIEAVTDALRDEV
MUL_3423|M.ulcerans_Agy99 VLPDMRARRSG-AIVNISSIAVALTPVGSGYYAAAKAAMEGMSGALHGEL
Mb3199|M.bovis_AF2122/97 FADRIAERSG--AIVNVSSVLAWLPLGMS--YGVSKAAMWSATESMRIEL
Rv3174|M.tuberculosis_H37Rv FADRIAERSG--AIVNVSSVLAWLPLGMS--YGVSKAAMWSATESMRIEL
Mvan_0234|M.vanbaalenii_PYR-1 FVPVIERNGGG-AILNVLSVLSWLHPPTSGAYSAAKAAGWAMTDALRAEL
MSMEG_1595|M.smegmatis_MC2_155 FAPVLADNGGG-AFLNVLSAAIWIPVPTA--YAASKAAAWSATNAMRAEL
MAB_4428|M.abscessus_ATCC_1997 FAPILKANGGG-AIVNIHSVLSWLGGAGA--YGASKAAIWSVSNTLRLEL
MLBr_00862|M.leprae_Br4923 FGQRLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMFSDCLRAEL
TH_3037|M.thermoresistible__bu VYKKMAKRGGG-AIVNQSSTAAWLY---SNYYGLAKAGLNSLTQQLATEL
MMAR_0254|M.marinum_M FLPAMVERGRG-AVLNVASVAGFAPLPGQAAYGAAKAFVLSYTHSLRGEL
MAV_4667|M.avium_104 LLPKLVDSGDG-LIVTVTSVAALEVYDGGAGYTAAKHGQGALHRTLRGEL
. : .:. * * :* : *:
Mflv_0930|M.gilvum_PYR-GCK STHGISVLAVEPGAFRTNAYAGFANKPVAETIPSYHDMLEEVRAAFVEMD
MUL_3423|M.ulcerans_Agy99 APLGISVTVVEPGAFRT-DFAGRSLVQSTRMINDYAATAGQRRKENDTMH
Mb3199|M.bovis_AF2122/97 APRGVQVVGVYVGLVDTDMGRFADAP-----------KSDPADVVRQVLD
Rv3174|M.tuberculosis_H37Rv APRGVQVVGVYVGLVDTDMGRFADAP-----------KSDPADVVRQVLD
Mvan_0234|M.vanbaalenii_PYR-1 APRGIHVAALHVGFMDTDMVSYIPADQ----------KTDPAVVAELALD
MSMEG_1595|M.smegmatis_MC2_155 TAQGTTVTAVVVGMVDTAMSARWDMP-----------KVSAESVVEQSLD
MAB_4428|M.abscessus_ATCC_1997 EAQHTHVLGVHAAFIDTDMVSALPVP-----------KTPPSAIAARIID
MLBr_00862|M.leprae_Br4923 DAVGVGLTTICPGVIDTNIIQSTRIDTPTAK----QERVRDRRGQLDMMF
TH_3037|M.thermoresistible__bu GGQNIRVNAIAPGPIDTEANRSTTPK-------------EMVDDIVKRIP
MMAR_0254|M.marinum_M HGSGVSVTALSPGPVDTSFGDVAGFTQQETESLPKIMWKSVDQVAQAGID
MAV_4667|M.avium_104 LGKPVRLTEIAPGAVETEFSLVRFDGD----------QQRADAVYTGITP
: : . . *
Mflv_0930|M.gilvum_PYR-GCK GVQPGDPRRGARAVIAAMAQDPPPRRLVLGNSGYDAVIDALEQSLGDIRA
MUL_3423|M.ulcerans_Agy99 GNQPGDPAKAAAAIITAVESPAPPAFLLLGPDALMAYRHIAQRRDVEITD
Mb3199|M.bovis_AF2122/97 GIEAGKEDVLADEMSRQVRASLNVPARERIARLMGN--------------
Rv3174|M.tuberculosis_H37Rv GIEAGKEDVLADEMSRQVRASLNVPARERIARLMGN--------------
Mvan_0234|M.vanbaalenii_PYR-1 GLFAGQPEILADELTRNVKAQLSAAPA-----------------------
MSMEG_1595|M.smegmatis_MC2_155 GVLSGAFEVLADDDSRLAKSLLSEPYEELHAVAVQALSDFKP--------
MAB_4428|M.abscessus_ATCC_1997 ALDKGDNEVLTDDLTVQIKSQLAGPVEKLAYRPGH---------------
MLBr_00862|M.leprae_Br4923 RLRRYGPDKVADTILSAIKKNKPIRPVAPEAYALYGISRVLPQVLRSTAR
TH_3037|M.thermoresistible__bu LSRLGEPEDLVGMCLFLLSDQAKWITGQIFNVDGGQIIR-----------
MMAR_0254|M.marinum_M GLAAGKAVVVPGLANRISAELIRVTPPDWMGSIMARIHPAMKRQ------
MAV_4667|M.avium_104 LVAADVAEVIGFVASRPAHVNLDLIVMKPRDQASASRFNRRG--------
Mflv_0930|M.gilvum_PYR-GCK NETLSRSADFPT
MUL_3423|M.ulcerans_Agy99 WEQLTAGTNLTP
Mb3199|M.bovis_AF2122/97 ------------
Rv3174|M.tuberculosis_H37Rv ------------
Mvan_0234|M.vanbaalenii_PYR-1 ------------
MSMEG_1595|M.smegmatis_MC2_155 ------------
MAB_4428|M.abscessus_ATCC_1997 ------------
MLBr_00862|M.leprae_Br4923 IRVI--------
TH_3037|M.thermoresistible__bu ------------
MMAR_0254|M.marinum_M ------------
MAV_4667|M.avium_104 ------------