For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGPYRQGIAERRKESWTSPDTATDSGGMNLKNARVLLVGGTSGIGLGVATAVAERGGVPIVASRNQASVD RALGTLPDGARGLTVDLTDPAALAGFAAQVGPIEHLVFTAGEELQMYPLADITAEAIGDLFGTRFAGQLL SVRAFAPRITAGGSITLTSGTAADKPDLGVLPAAVCGAVSAMTRALAVELAPLRVNAVAPGPIDTPLWSA LAPEGRDALYAQYAQQLPAGRMGAVTDTALAYTYLMEQEFGTGQVLTVDGGGTLV
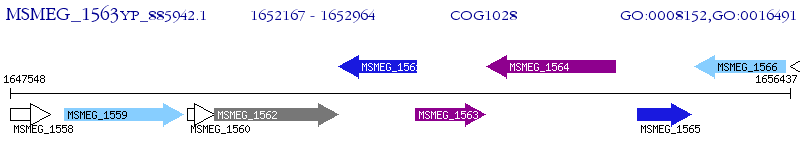
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1563 | - | - | 100% (265) | short-chain dehydrogenase/reductase SDR |
| M. smegmatis MC2 155 | MSMEG_6823 | - | 2e-23 | 33.20% (241) | short-chain dehydrogenase/reductase SDR |
| M. smegmatis MC2 155 | MSMEG_3367 | - | 3e-22 | 32.92% (243) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2788c | fabG | 3e-15 | 27.35% (245) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_2071 | - | 3e-20 | 30.93% (236) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2766c | fabG | 3e-15 | 27.35% (245) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 1e-14 | 30.49% (246) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_0057 | - | 2e-26 | 32.92% (240) | putative short chain dehydrogenase/reductase |
| M. marinum M | MMAR_0587 | - | 2e-28 | 35.54% (242) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_2775 | - | 3e-18 | 31.08% (251) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_2488 | - | 3e-17 | 32.26% (248) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_0315 | fabG2 | 3e-18 | 30.80% (237) | 3-oxoacyl- |
| M. vanbaalenii PYR-1 | Mvan_0325 | - | 9e-20 | 32.31% (260) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
TH_2488|M.thermoresistible__bu ---------------------------MEFQGLHVLVTGGTGGIGLACAR
Mvan_0325|M.vanbaalenii_PYR-1 ----------------------MNTTAATLSGRTALVTGSTSGLGAGIAE
Mb2788c|M.bovis_AF2122/97 ------------------------MTSLDLTGRTAIITGASRGIGLAIAQ
Rv2766c|M.tuberculosis_H37Rv ------------------------MTSLDLTGRTAIITGASRGIGLAIAQ
MAV_2775|M.avium_104 -----------------------MLDLFDLRGRRALVTGASTGIGKQVAQ
MLBr_01807|M.leprae_Br4923 -----------MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVAR
MUL_0315|M.ulcerans_Agy99 --------------------MRPD-SSPSWCTITSVVTGGASGIGLAIAR
Mflv_2071|M.gilvum_PYR-GCK --------------------------MSNSSERVAVIVGGASGIGWATAQ
MMAR_0587|M.marinum_M --------------------------MGQLEGKTALVTGGNSGIGLAAAQ
MSMEG_1563|M.smegmatis_MC2_155 MGPYRQGIAERRKESWTSPDTATDSGGMNLKNARVLLVGGTSGIGLGVAT
MAB_0057|M.abscessus_ATCC_1997 --------------------MTGFLSGTAFADRTVVLIGGGTGIGLSVAR
:: *. *:* *
TH_2488|M.thermoresistible__bu AFAEAGAEVTVTGRDA-GRG----AAAAGAHPRLRFIRGDLAD-LASVQN
Mvan_0325|M.vanbaalenii_PYR-1 ALADAGAHVVITGRTR-ARGEQVVTQIESQGGRAVFVPVDFAAGPGTITD
Mb2788c|M.bovis_AF2122/97 QLAAAGAHVVLTAR----RQEAADEAAAQVGDRALGVGAHAVD--EDAAR
Rv2766c|M.tuberculosis_H37Rv QLAAAGAHVVLTAR----RQEAADEAAAQVGDRALGVGAHAVD--EDAAR
MAV_2775|M.avium_104 AYLQAGARVAIAARDFEALQRTADELTAGGTGGVVAIRCDVTQ--PDEVS
MLBr_01807|M.leprae_Br4923 RLVADGHRVAVTHRAS----V--------VPDWFFGVECDVTD--NDAIG
MUL_0315|M.ulcerans_Agy99 RLSADGTDVATLDLK--------------TPDGGLAFSADVTN--RSQVD
Mflv_2071|M.gilvum_PYR-GCK ALAAEGCRVTIADRNA----DGARERASELGSGHRAEAVDVTD--EDSVA
MMAR_0587|M.marinum_M RLAAEGAHVFLTGRNQ----ATIDAAVASIGSRAHGIRADVSN--IEDLA
MSMEG_1563|M.smegmatis_MC2_155 AVAERGGVPIVASRNQ----ASVDRALGTLPDGARGLTVDLTD------P
MAB_0057|M.abscessus_ATCC_1997 LVTAAGGDVVLGGRTP----ATLAVAAEELGGKARWHLVDTAH------Q
* .
TH_2488|M.thermoresistible__bu LIR-----YADAVDVLVNNAASFPG-ALTVEQDAESFTRTFATNVTGTYF
Mvan_0325|M.vanbaalenii_PYR-1 LVVTATEALGGRLDILVNNVATLVQPAPTADVTAEEITTAFGVSVATPFL
Mb2788c|M.bovis_AF2122/97 RCVDLTLERFGSVDILINNAGTNPAYGPLLEQDHARFAKIFDVNLWAPLM
Rv2766c|M.tuberculosis_H37Rv RCVDLTLERFGSVDILINNAGTNPAYGPLLEQDHARFAKIFDVNLWAPLM
MAV_2775|M.avium_104 GMVDRMIAELGGIDIAVCNAGIISVS-PMLEMPAAEFQRIQDTNVTGVFL
MLBr_01807|M.leprae_Br4923 AAFKAVEEHQGPVEVLVANAGITSDM-LIMRMSEEQFQKVIDVNLTGVFR
MUL_0315|M.ulcerans_Agy99 AALAAIRQRLGPVTILVNAAGLDGFK-RFSNITFEDWQRVIDVNLNGVFH
Mflv_2071|M.gilvum_PYR-GCK RLFDAI----GPLDIAVNCAGFSNVG-LITDMAAEDFRAVIDVCLNGAFI
MMAR_0587|M.marinum_M RVADAIAAAGRGLDVLFANAG-GGEFVLLGDITIEHFTNGFMTNVAGTLF
MSMEG_1563|M.smegmatis_MC2_155 AALAGFAAQVGPIEHLVFTAGEELQMYPLADITAEAIGDLFGTRFAGQLL
MAB_0057|M.abscessus_ATCC_1997 ASIDAFFDQI-EVLHGLFTTAATYVTGSIAALSVEQAATPFESKFWGQYR
: . .. .
TH_2488|M.thermoresistible__bu LTAGLIPGMLARGRGSIVNITSMVAT---KAVPGASVYSSSKAAVESLTR
Mvan_0325|M.vanbaalenii_PYR-1 LTGLIAPAMVQRGDGAIVNIGSISGI---IGAAGSALYGATKASVHLLTK
Mb2788c|M.bovis_AF2122/97 WTSLVVTAWMG-EHGGAVVNTASIGG--MHQSPAMGMYNATKAALIHVTK
Rv2766c|M.tuberculosis_H37Rv WTSLVVTAWMG-EHGGAVVNTASIGG--MHQSPAMGMYNATKAALIHVTK
MAV_2775|M.avium_104 TAQAAARAMVRQGRGGAIITTASMSGHIINVPQQVGHYCASKAAVIQLTK
MLBr_01807|M.leprae_Br4923 VAKLASRNMIKQRFGRYIFMGSVVGL---SGVPGQANYAASKSGLIGMAR
MUL_0315|M.ulcerans_Agy99 TIQAVLPDMVAAGWGRIVNISSSSTH---SGTPYMTHYVAAKSAVNGLTK
Mflv_2071|M.gilvum_PYR-GCK VTKHAGRHLRRG--GSLVQISSLNGR---QPAAGMSAYCAAKAGLSMLTQ
MMAR_0587|M.marinum_M TVQTMLPLLNSG--ASIVLTGSTAAY---NGSPAFSVYAATKAAIRSFGR
MSMEG_1563|M.smegmatis_MC2_155 SVRAFAPRITAGGSITLTSGTAADKPD----LGVLPAAVCG--AVSAMTR
MAB_0057|M.abscessus_ATCC_1997 VVKSALPKLSSDASVVLMSGAASVRP-----AGSAPAYVAANAAIEGLAR
: . .: . :
TH_2488|M.thermoresistible__bu SWAVEFAAHGIRVNAVAPGPTSTDGVATEWGETN-----EALGRALPIGR
Mvan_0325|M.vanbaalenii_PYR-1 AWAAEYGPSGVRVNAVAPGPIATERTR-EFGDAI-----APVLARVPSHR
Mb2788c|M.bovis_AF2122/97 QLALELSPR-IRVNAICPGVVRTRLAEALWKDHED-----PLAATIALGR
Rv2766c|M.tuberculosis_H37Rv QLALELSPR-IRVNAICPGVVRTRLAEALWKDHED-----PLAATIALGR
MAV_2775|M.avium_104 AMAVEFAPHDIRVNSVSPGYIRTELVEPLHEYHR------QWQPKIPLGR
MLBr_01807|M.leprae_Br4923 SLARELGSRSITANVVAPGFIDTEMTRAMDSK-----YQEAALQAIPLGR
MUL_0315|M.ulcerans_Agy99 ALALEYGPSGITVNAVPPGFIDTPMLRNAAAQGHLG-DIEQNIARTPVRR
Mflv_2071|M.gilvum_PYR-GCK VAALEMGPRGIRVNAVAPGFVHTPLTAAAASVP--G-VVEDYVDNTALGR
MMAR_0587|M.marinum_M TWAAELVSRNIRVNTVVPGPVETPGLKGLAPEGQEQQLLEGEAAKVPMGR
MSMEG_1563|M.smegmatis_MC2_155 ALAVELAPL--RVNAVAPGPIDTPLWSALAPEGRD-ALYAQYAQQLPAGR
MAB_0057|M.abscessus_ATCC_1997 GLAHELAPI--RVNALSPGTVDGHLWNQRPPEIRR-QAFEHIAAAATLRR
* * . .* : ** . *
TH_2488|M.thermoresistible__bu TADPDEIAQAVVFLASSRASFITGAVLHVDGGGIAV--------------
Mvan_0325|M.vanbaalenii_PYR-1 MSTVAEVASAVVFLAGPEAANIHGTILSVDGGATAVRDEPAAVTKLGNHP
Mb2788c|M.bovis_AF2122/97 IGEPADIASAVAFLVSDAASWITGETMIIDGGLLLGNALGFRAAPSTEH-
Rv2766c|M.tuberculosis_H37Rv IGEPADIASAVAFLVSDAASWITGETMIIDGGLLLGNALGFRAAPSTEH-
MAV_2775|M.avium_104 MGRPDELAGIYLYLASAASSYMTGSDIVIDG--------GYSCP------
MLBr_01807|M.leprae_Br4923 IGAVEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH-------------
MUL_0315|M.ulcerans_Agy99 IGKPEDIAAACAFLVSEEAGYITGQILGVNGGRNT---------------
Mflv_2071|M.gilvum_PYR-GCK AGTPEDIAAAVVFLCSPGTSWMTGEVLDINGGAHLKRYPDVMGHVMKLME
MMAR_0587|M.marinum_M LGKPAEIAAAVLFLASDQSSFMTGSEMFVDGGTEQL--------------
MSMEG_1563|M.smegmatis_MC2_155 MGAVTDTALAYTYLMEQ--EFGTGQVLTVDGGGTLV--------------
MAB_0057|M.abscessus_ATCC_1997 PVTEGEVAQAAVYLLLN--SATTGSTLYTDGGYALR--------------
: * :* * : :*
TH_2488|M.thermoresistible__bu ---
Mvan_0325|M.vanbaalenii_PYR-1 HPG
Mb2788c|M.bovis_AF2122/97 ---
Rv2766c|M.tuberculosis_H37Rv ---
MAV_2775|M.avium_104 ---
MLBr_01807|M.leprae_Br4923 ---
MUL_0315|M.ulcerans_Agy99 ---
Mflv_2071|M.gilvum_PYR-GCK PAS
MMAR_0587|M.marinum_M ---
MSMEG_1563|M.smegmatis_MC2_155 ---
MAB_0057|M.abscessus_ATCC_1997 ---